9) Index the crystal using the fast scan
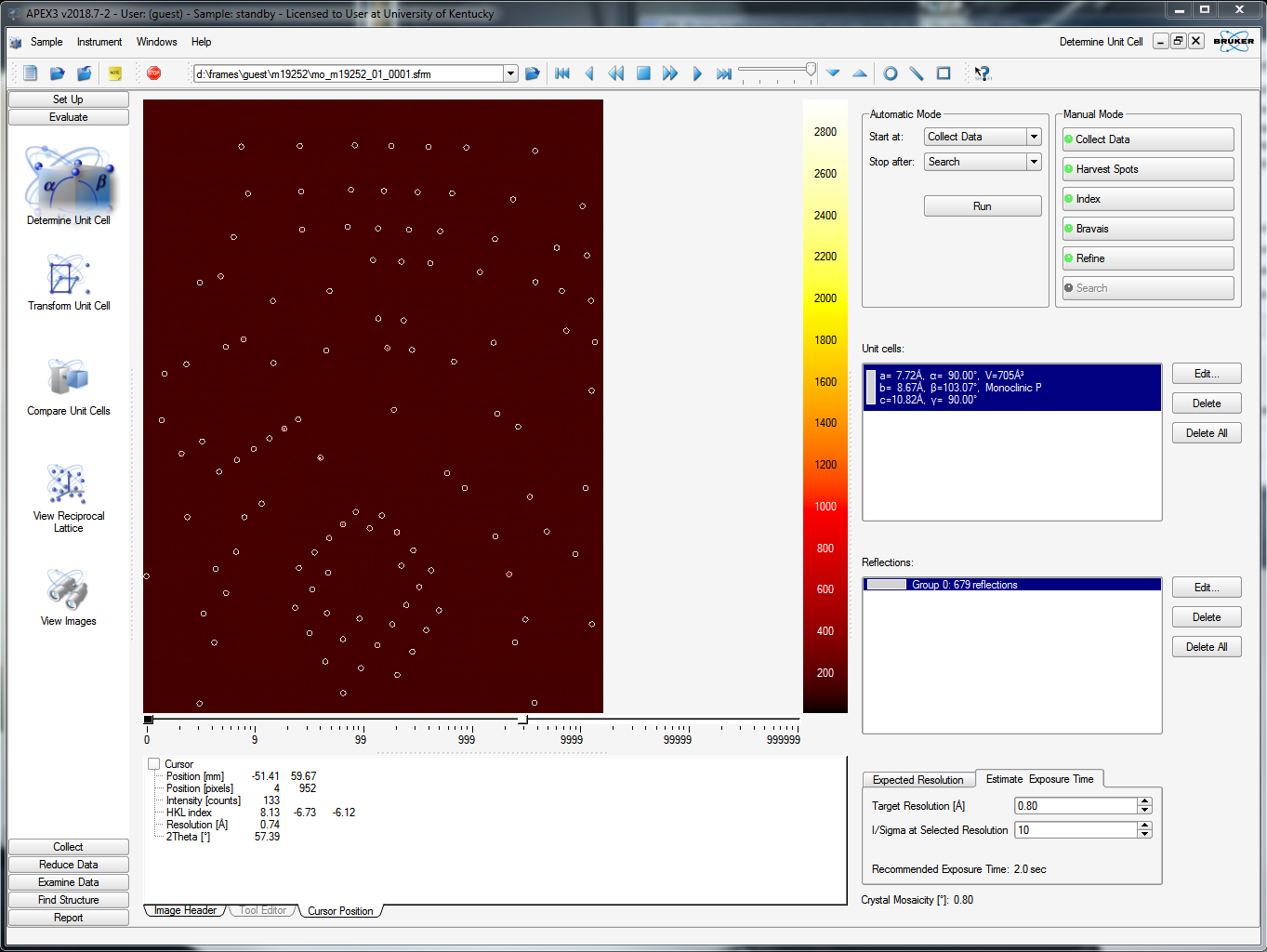
Once the fast scan is running and you've watched a few frames whiz by on the APEX3 window, you should try to index the crystal. On the left-hand-side vertical menu, select "Evaluate" and then "Determine Unit cell". The default APEX3 window for this functionality is shown below. In the upper right of the window, find the "Number of Runs:" box - it should be set to 1. That's your fast scan! Next to it there's a box for "Images per Run:". The default entry here is 20, but you should set it to the number of fast-scan frames that have already been collected. The more frames the better. In the image below it has been set to 60, which is plenty. Go ahead and click "Harvest" down in the bottom right.
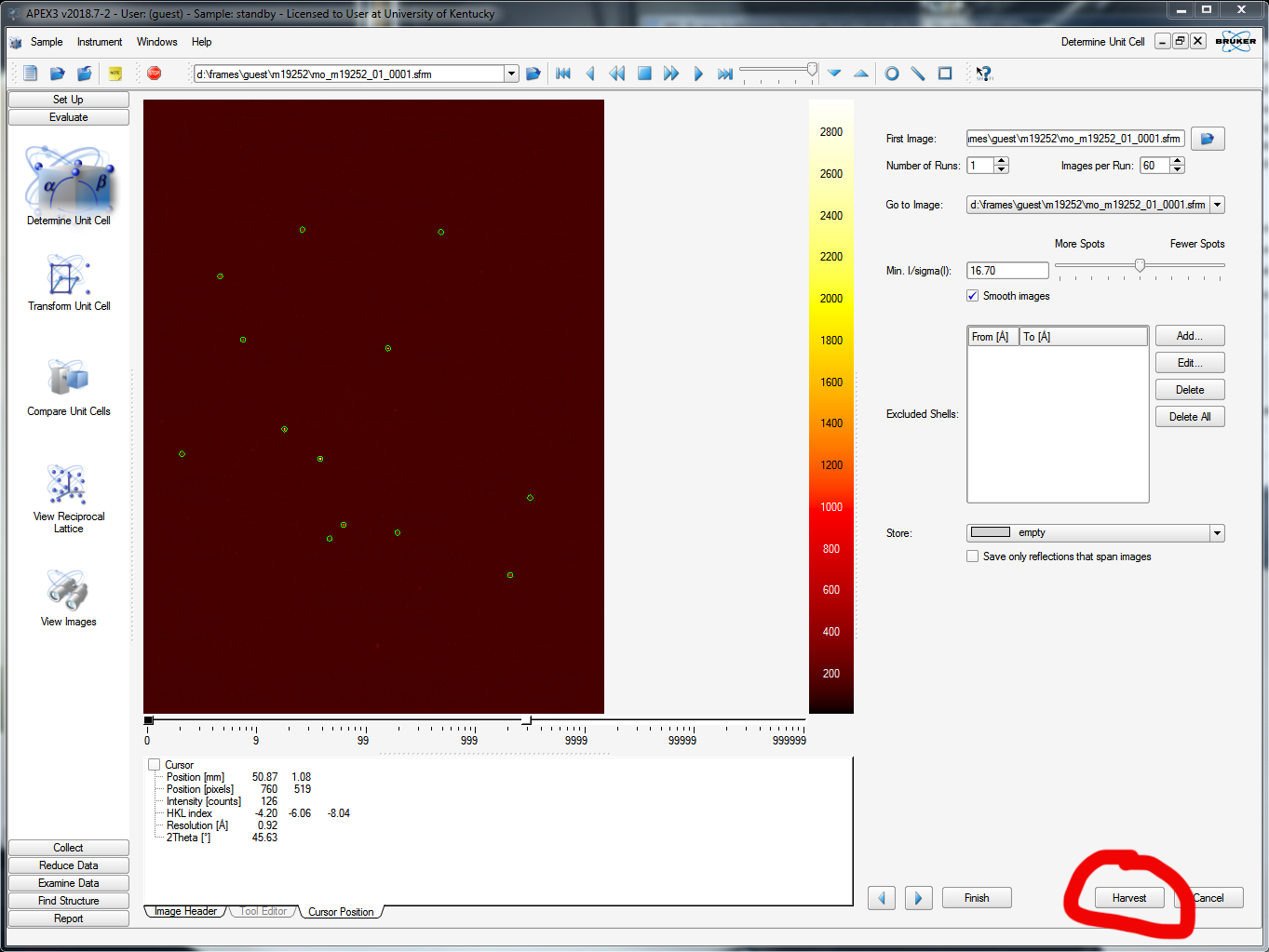
APEX3 scans through the specified number of images and finds the diffraction maxima in each image according to threshold criteria (see the slider in the above picture, upper right - about half way is a reasonable starting point, it should be obvious what it does). It saves the x and y coordinates and the rotation angle (cylindrical coordinates!). In this case it tells you it found 679 reflections, like this:
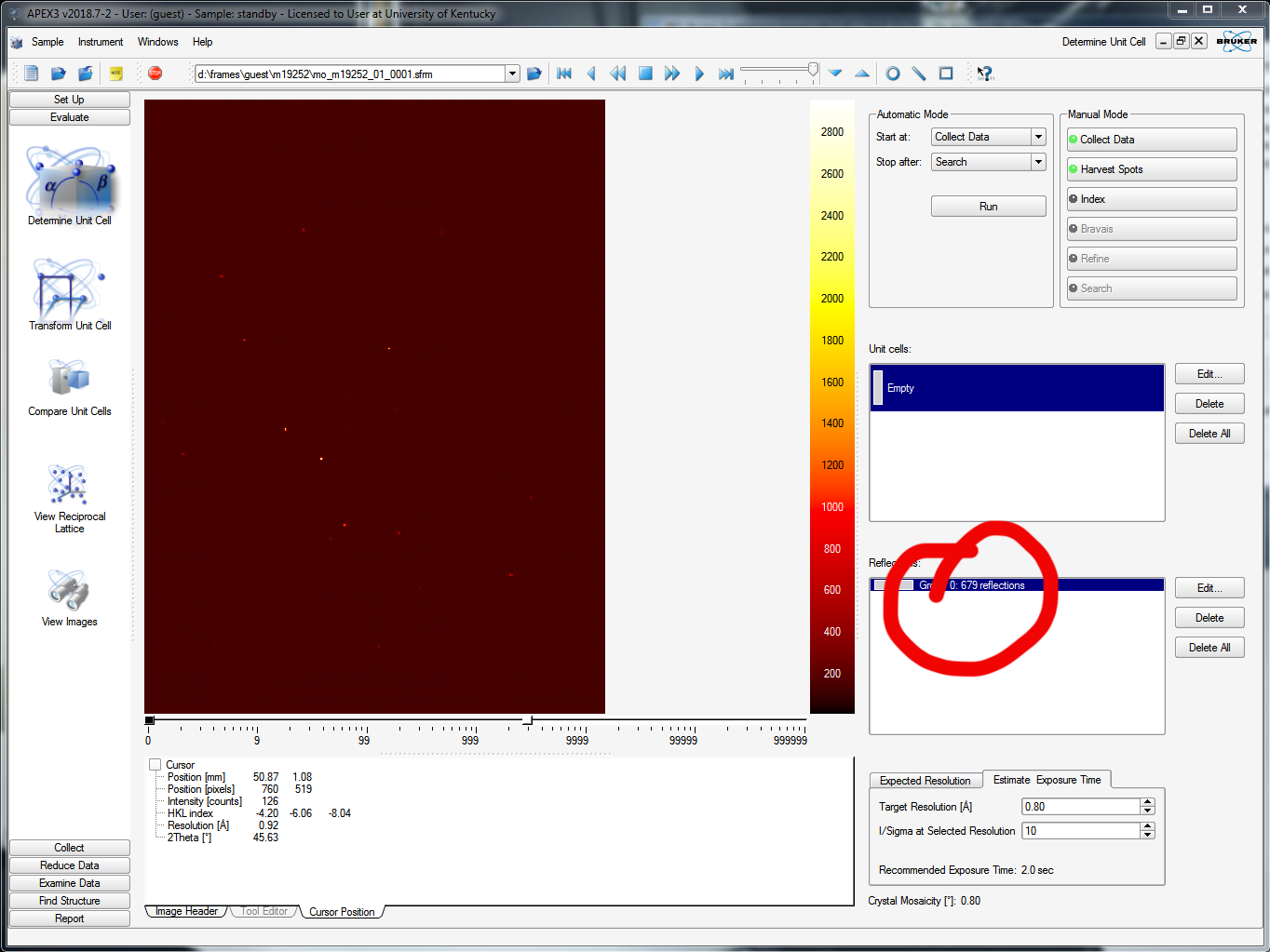
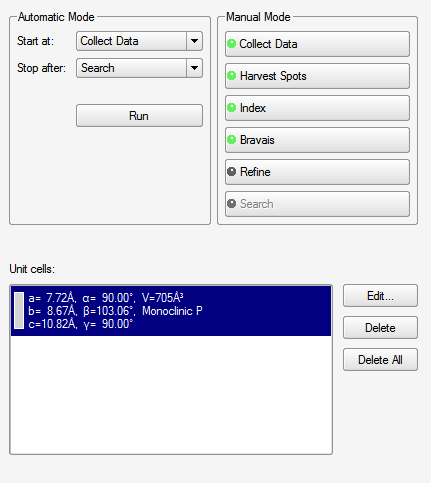
When you click "Index" you get the following:
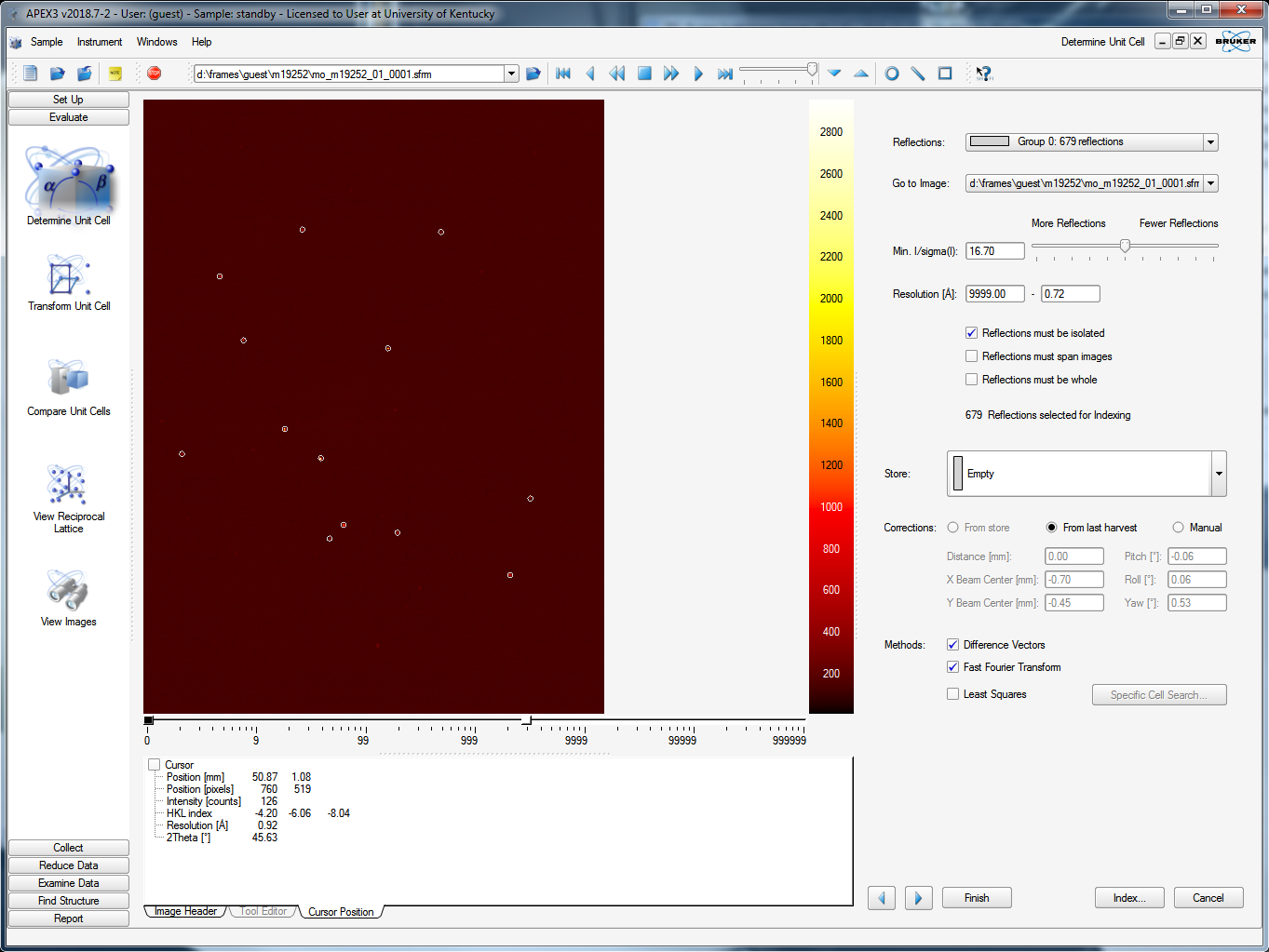
This allows you to fiddle a bit more with the reflections that will be used to index. On the first pass, there's little point in changing anything, so go ahead and click the "Index" button on the lower right. It tries two different algorithms: "difference vectors" and "fast Fourier transform". When both methods work (a good sign!) you get something like this snippet from the APEX3 window:
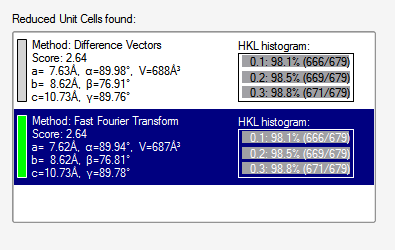
Sometimes one routine works while the other fails. Occasionally, if the crystal has some nasty problem, both will fail. There is a third indexing option, but it is beyond the scope of this exercise. Since both methods gave the same result, we could accept either. Sometimes they look a bit different but are equivalent, e.g., the β angle for monoclinic could be given as either acute or obtuse. When you accept one of the results, you get something like this (snippet):
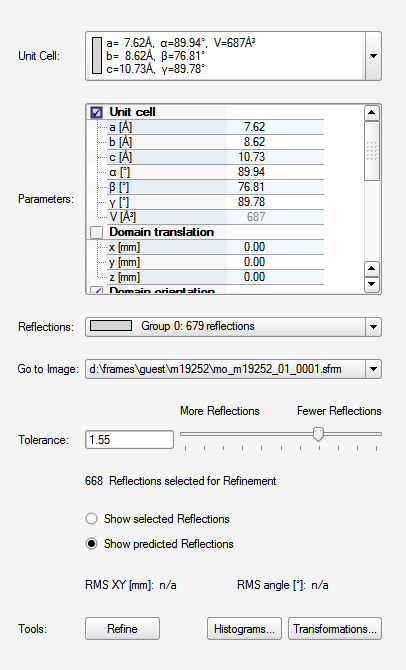
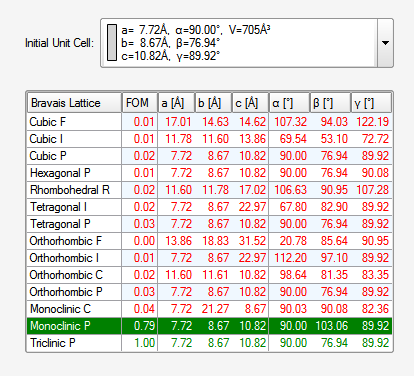
At this point you could click "Refine" to optimize the result a bit prior to transforming the unit cell to a standard setting and checking for higher symmetry. The goal is to find the standard Bravais lattice. You guessed it, click "Bravais" ...
... which brings up this table: