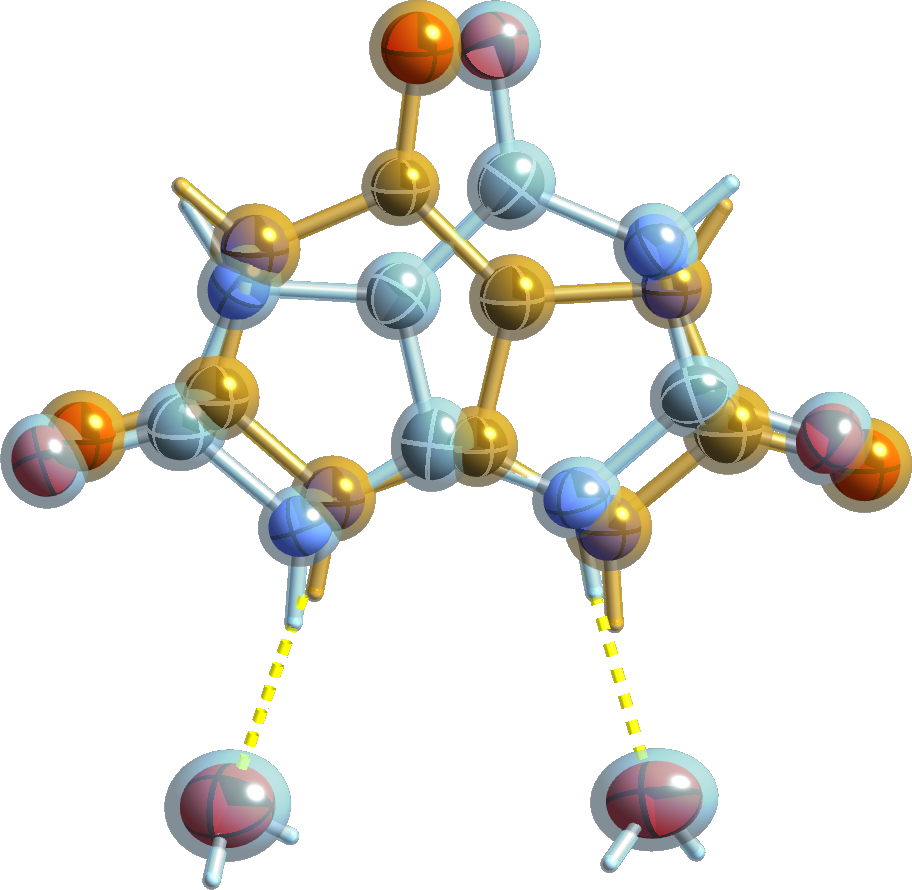
5/5) Tie the major/minor disorder components using FVARs
The data for this structure were collected many years ago (1990) on a serial diffractometer (Siemens P4 on a MacScience rotating anode). That instrument had a severe limitation: its maximum achievable 2θ angle of about 108.5°, restricted the attainable resolution to only 0.95Å with CuKα X-rays. As such, the data-to-parameter ratio is sub-optimal. Since the model requires whole-molecule disorder for the uric acid molecule, it must include coordinates and displacement parameters for two components. For this particular structure it is, however, possible to use just a single set of coordinates for both disorder components. The trick involves using FVAR parameters and careful manipulation of how SHELXL applies them to each fragment. Similarly, a single set of anisotropic displacement parameters may be used for both PARTs. When applied correctly, the two components become equivalent about the 2-fold rotation. Even the tilt and degree of eccentricity of the ellipsoids make sense. (Note: This only works because the β angle is so close to 90° (i.e., 90.51°); any distortion would be tiny because sin(90.51) = 0.99996, which is indistinguishable from unity for all practical purposes.)Setting up and applying the FVARs is not entirely trivial, and I'm cruel enough that I won't explain the setup process in detail. You can, however, download a *.res file to inspect. You should be able to figure the process out from there (and by reading the SHELXL manual). The end result looks like this: