
Unlike most structure solution programs, SHELXT does not need to know
the space group a priori. It works by first finding an expansion of the
structure using space group P1 and then tries to assign the proper space
group using the phases obtained for the P1 solution. In order to figure
out the space group, however, it does need to know the Laue group. In the above
file, LATT and SYMM indicate a primitive centrosymmetric monoclinic
unit cell. Once the structure is solved, SHELXT will insert the required
symmetry for P21/c as SYMM instructions.
SHELXT can be run from a command line or from the APEX3 GUI. For uric acid dihydrate, SHELXT gives three separate 'solutions', distinguished by the suffixes '_a', '_b', '_c' for space groups P21/c, P21, and Pc. Results for the P21/c structure are in file sp02_a.res, which looks like this:
SHELXT can be run from a command line or from the APEX3 GUI. For uric acid dihydrate, SHELXT gives three separate 'solutions', distinguished by the suffixes '_a', '_b', '_c' for space groups P21/c, P21, and Pc. Results for the P21/c structure are in file sp02_a.res, which looks like this:

Notice that the SYMM line now includes translations of a half for
y and z, which are required for P21/c.
Some diagnostics from the SHELXT run are also included on REM
lines. At this stage, all of the atoms of the uric acid molecule and the two
waters should be present. Although SHELXT tries to assign atom types
based on elements present on the SFAC line and the amount of electron
density it found, it does not always get all atoms correct, so a few of them
might be mis-assigned. As with any other structure, we should fix any errors
before moving on. If we load the structure into ShelXle, we should see
something like this:
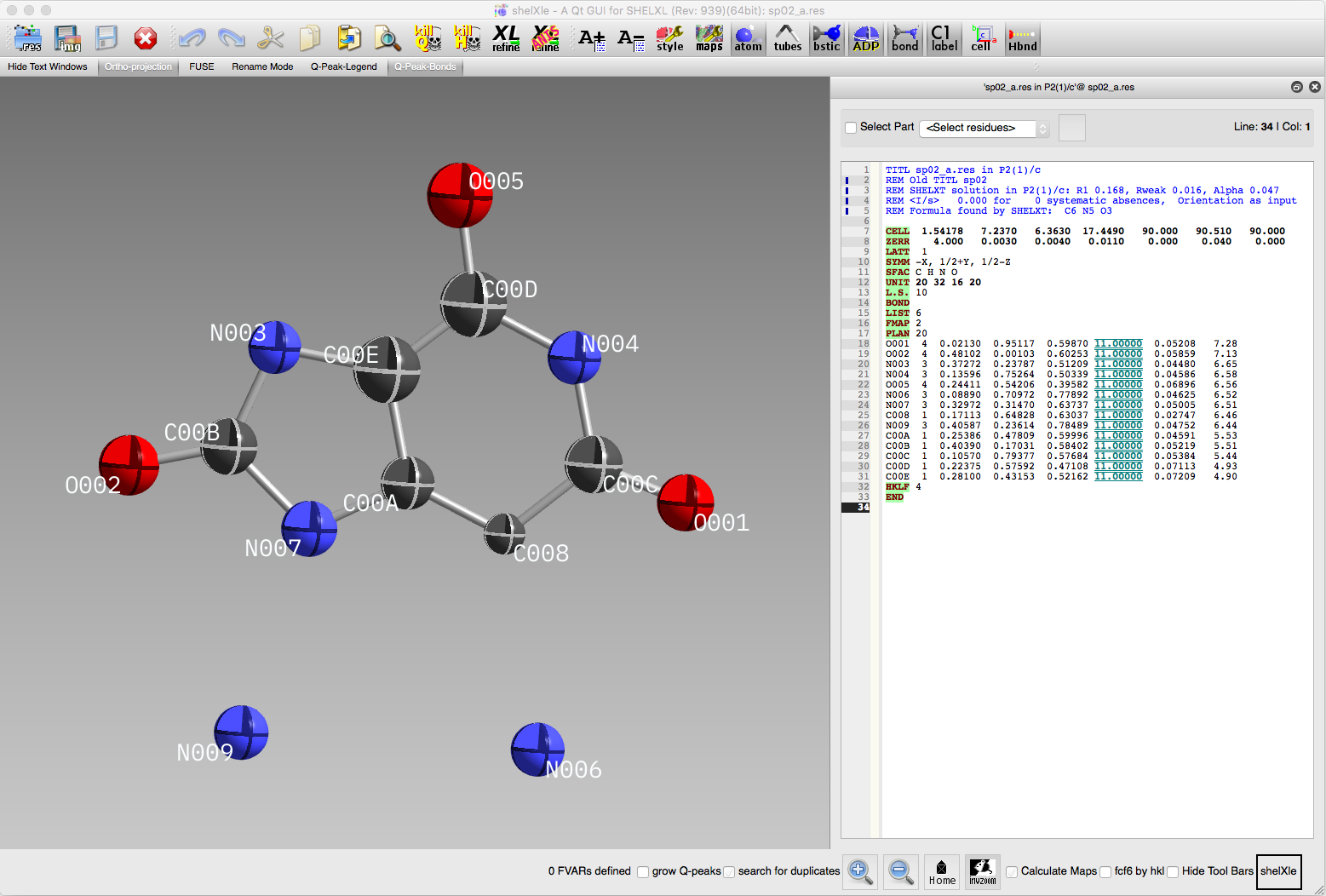
In the above ShelXle window, the two water molecule oxygen atoms were
assigned by SHELXT to be nitrogen, and the atom labelled C008 ought to
be nitrogen, not carbon. These are easily fixed in ShelXle. For example,
the right mouse button brings up several options, one of which is "Change
element to...". For this exercise, however, we'll jump straight into "Rename
Mode" so that we can fix atom types and assign a sensible numbering scheme
all at one go. We might as well also use ShelXle's SORT routine
(it's in the SHELX dropdown menu). The result ought to look something
like this:
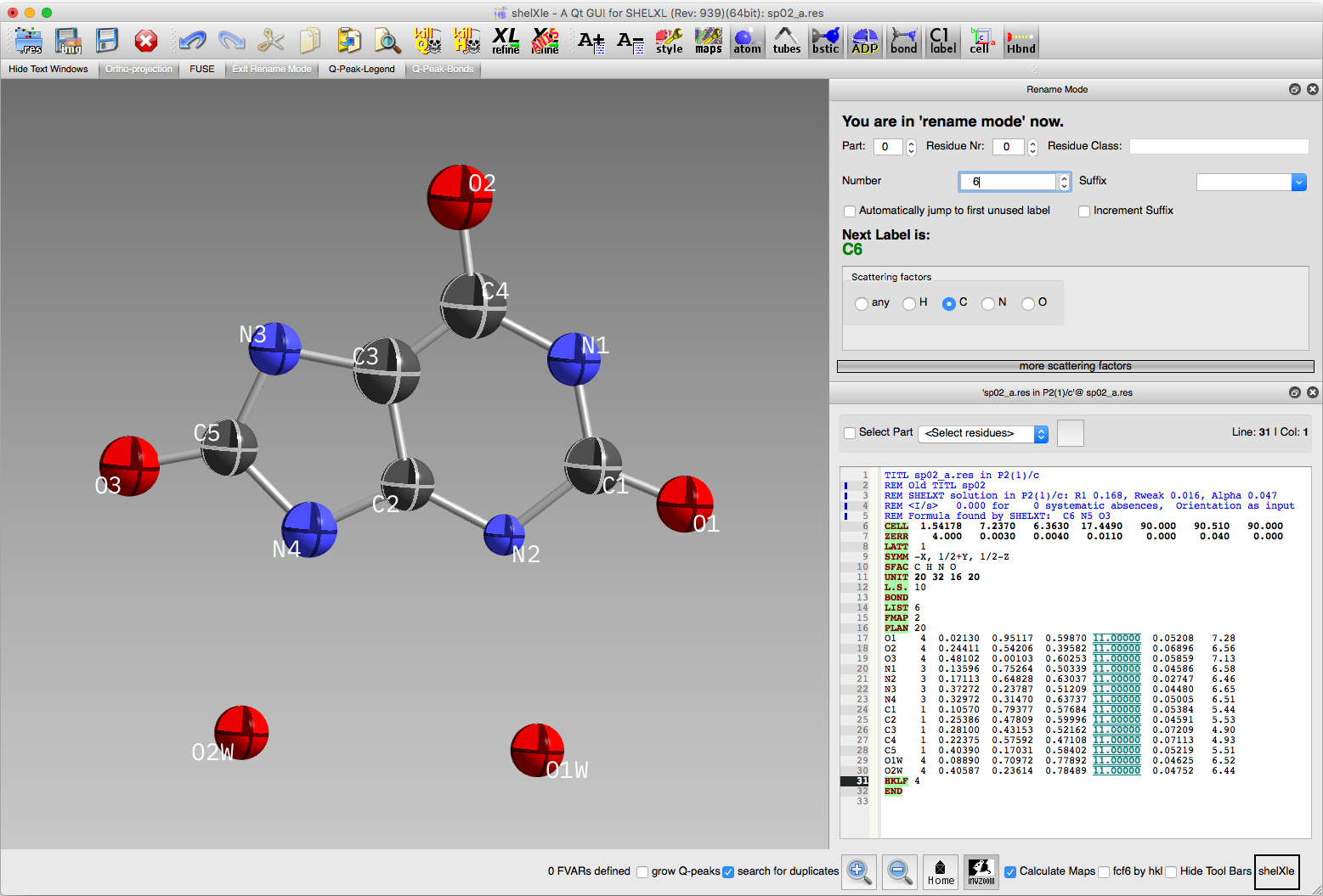
It is always best practice to assign a sensible numbering scheme and to
sort the list of atoms into a logical order. The ShelXle text editor on
the right side of the above image shows the current SHELXL *.ins/*.res
file.
In the next section we will account for twinning. In preparation for that,
we'll run a few cycles of least-squares refinement with SHELXL, which
gives us an idea of how lousy the fit is prior to modelling twinning and
disorder. The result should look something like the following:
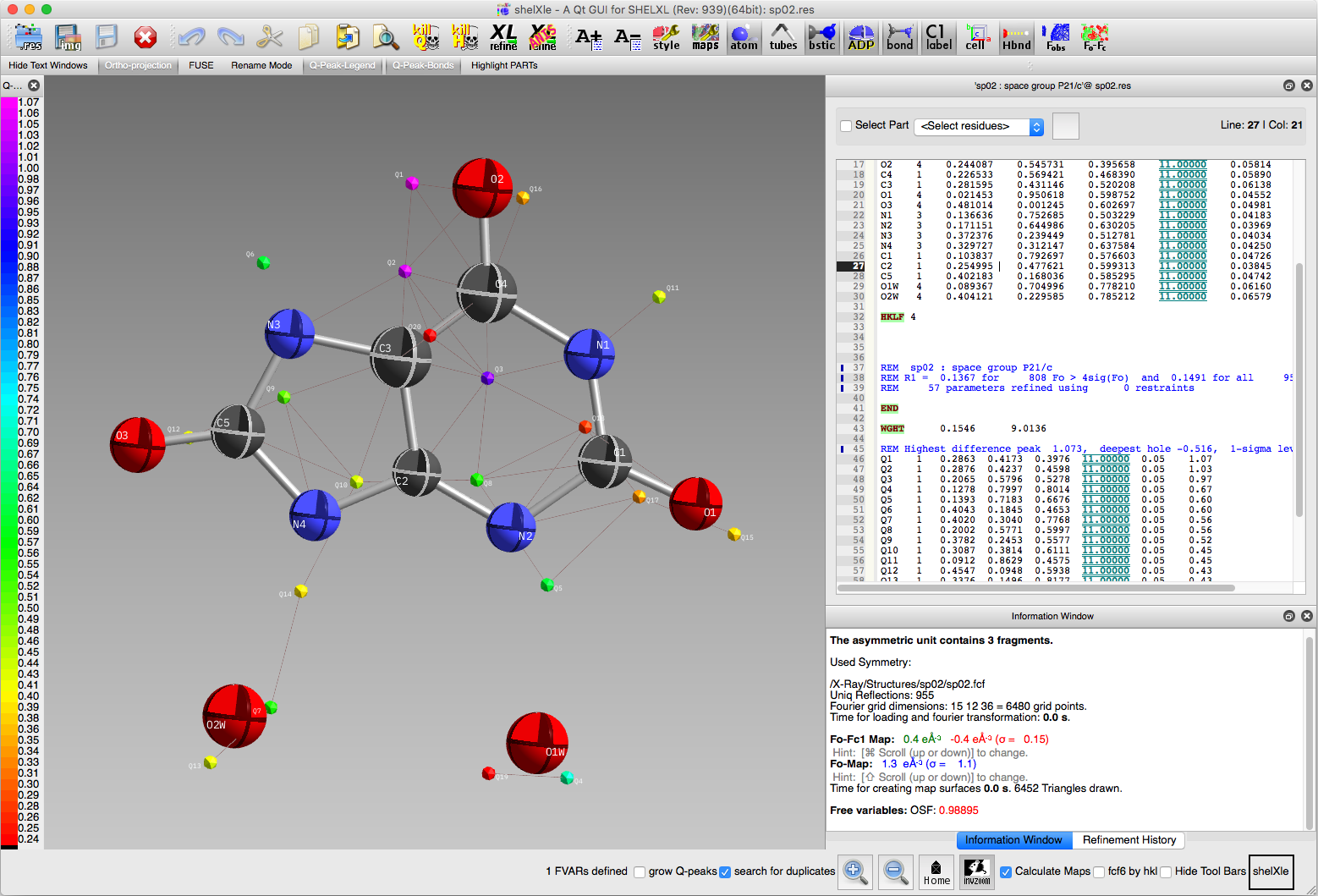
The ShelXle display also shows some difference map peaks. The largest
of these are likely candidates for disordered atoms, but we'll ignore them
for now. In section 2, we'll deal with
the twinning problem.
1) Solve structure using SHELXT and ShelXle.
2) Include twinning using TWIN and BASF in SHELXL.
3) Add disorder using FRAG...FEND in SHELXL.
4) Add restraints/constraints to enable stable refinement.
... More advanced stuff (only possible because β is very close to 90°)...
5) Tie the major/minor disorder components using FVARs.
2) Include twinning using TWIN and BASF in SHELXL.
3) Add disorder using FRAG...FEND in SHELXL.
4) Add restraints/constraints to enable stable refinement.
... More advanced stuff (only possible because β is very close to 90°)...
5) Tie the major/minor disorder components using FVARs.
Return to the first page of this tutorial
Return to the main Tutorials page or to the main X-Ray Lab page
Return to the main Tutorials page or to the main X-Ray Lab page