1/7: Introduction
A drawback of retirement is lack of easy access to the full version of Mercury from the CCDC. One particular feature absent in the free version is creation of least-squares-fit superpositions of related structures, so I decided to develop my own. The process of making structure overlays in Mercury is (for me) overly fiddly, so I went about it in a different way. It requires some diligence in preparation, but fits in better with my workflow. The procedure, programs, and scripts are described herein. An example for a simple two-structure alignment follows.
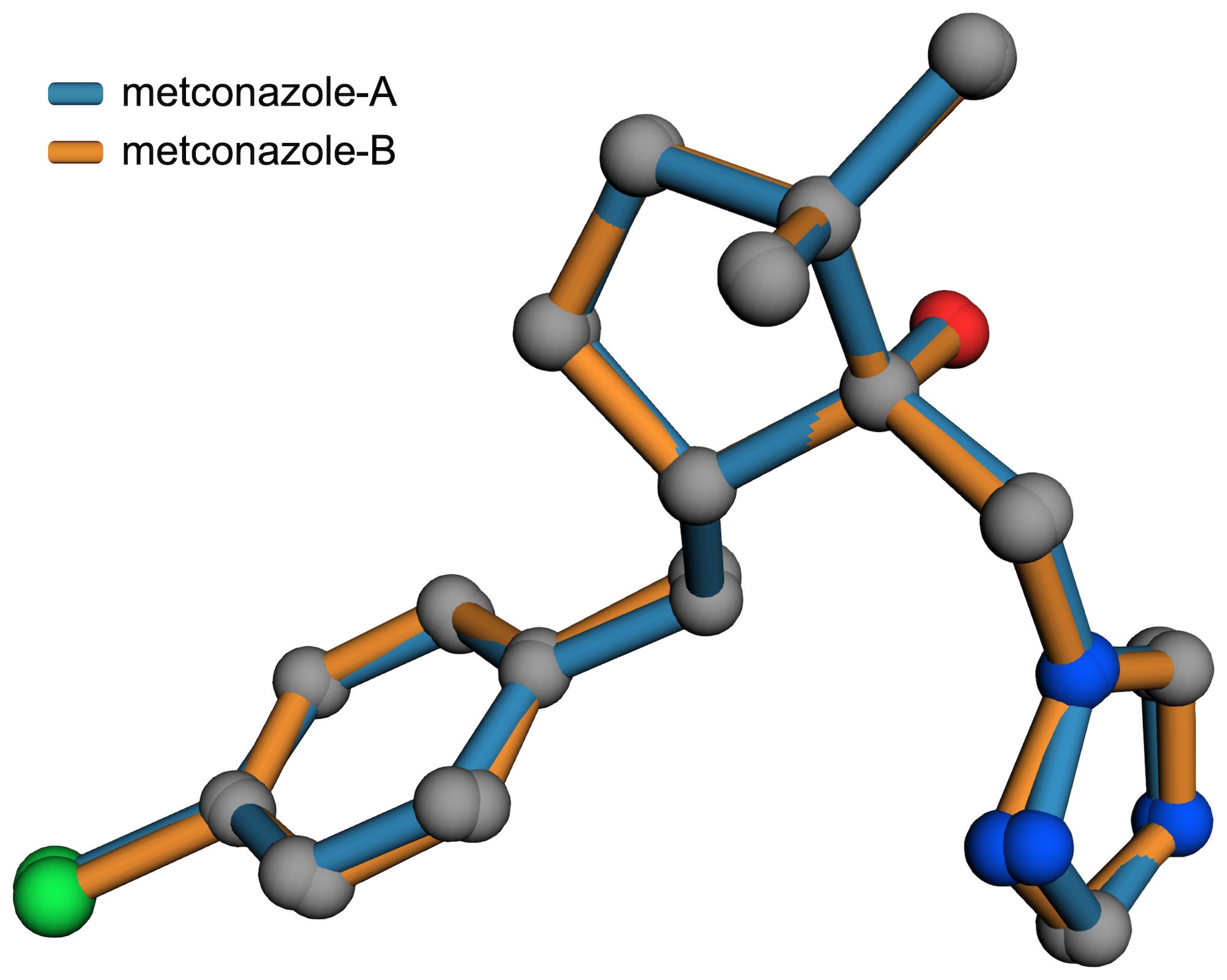
This particular structure is of the agricultural fungicide metconazole, which has two similar conformers in its asymmetric unit. We'll use it as an example covering how to prepare input files for the superposition and for creating high-quality, high-resolution figures. Incidentally, the overlay plot in the metconazole paper was drawn with Mercury, so you can easily compare with the above figure.
We'll be using Python, JavaScript, plus a bunch of plug-in modules for each etc. Hopefully all the requirements are explained in enough detail that it's painless to implement. The programs/scripts were written on a Mac flaptop, but should be easy to make run on Linux. I don't use Windows, but it ought to work with minor changes. One of the great things about Python and JavaScript is that somebody, somewhere has almost certainly written code that does all the seemingly difficult stuff. Moreover, AI can be phenomenally good at de-bugging, suggesting, and even implementing new features. DeepSeek and Github Copilot were especially useful for the JavaScript code. All visual eye candy is done in a browser, so should be cross-platform no matter what.