6/6) KappaCCD: Final Merging with XPREP
The title is a bit of a misnomer, as the intention here is to create a combined,
fully scaled, but unmerged dataset. The reason to keep it unmerged is so that
SHELXL will do the merging on-the-fly, and thereby write sensible numbers for
Rint etc. in the CIF later on.
This assumes you are somewhat familiar with XPREP. Start the program and read
in the reference set. This is most likely going to be your edited 'slow' scan '.hkl'
file.
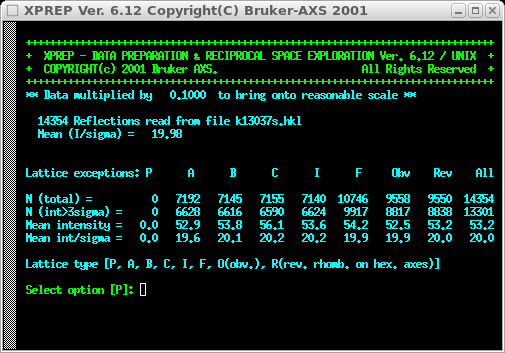
You need to go through the regular XPREP stuff to set lattice type, space
group etc. If you used makep4p earlier, and copied the 'smart.p4p'
file to the file name of your datasets, then XPREP will be able get some of
the needed information from the '.p4p' file. Not all of the input stuff is shown in
the images here, but at some point you will get the following screen:
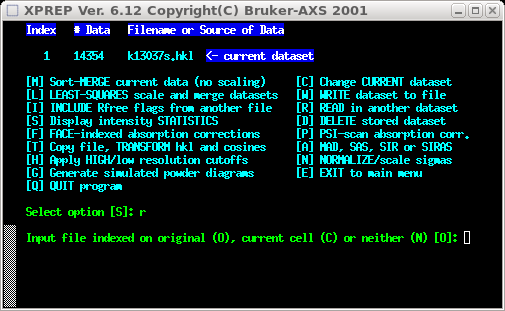
Select [R] to read in another dataset. You're going to read in the 'fast' set, so
select [O] because it was indexed in the same way.
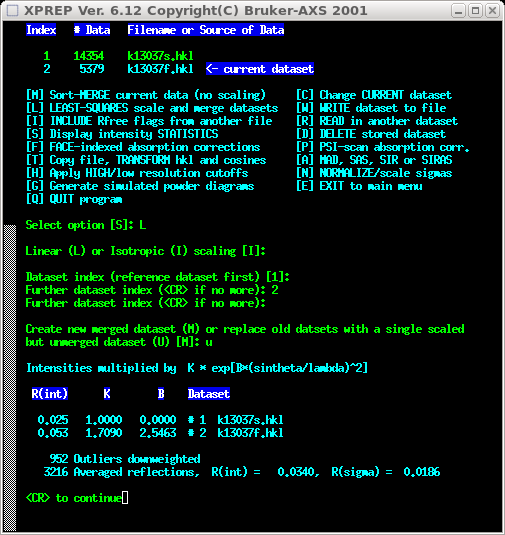
Again, a few keyboard inputs are skipped in the above picture. Select [L] to do
a least-squares scale and merge. In this case either linear or isotropic should
be fine (it's the same crystal, same temperature etc. so perhaps linear is
more 'correct' - many tests suggest that for this particular application it makes
little difference). You get the option to save a merged or unmerged set ... choose
unmerged [U].
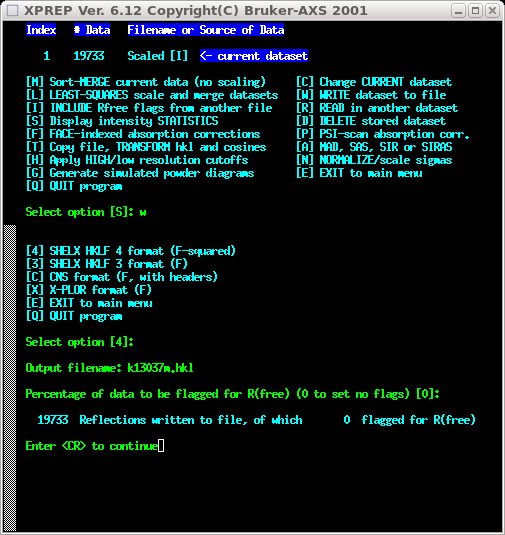
Lastly, you need to write out the combined, unmerged but fully scaled dataset.
Here we create an '.hkl' file with an 'm' designator to distinguish it from the
individual 'fast' and 'slow' files. You can of course name it however you want,
but remember to be consistent. You can safely ignore the R(free) stuff - it's
used for bias awareness in macromolecular crystallography and has little use
here (ask me and I'll be glad to explain).
You now have a scaled and combined '.hkl' file with the best treatment of overloaded
intensities that you are likely to get after integration by Denzo. Other
programs (e.g. EVAL15 and APEX2) may give comparable results,
but require a different sort of additional effort.
1) Extract the intensity data from the diffraction frames.
2) Run a first pass through with Scalepack.
3) Create '.sad' input files for SADABS.
4) Run SADABS separately for fast and slow scans.
5) Identify overload intensities and edit slow .hkl file.
6) Merge fast and slow scans in XPREP.
2) Run a first pass through with Scalepack.
3) Create '.sad' input files for SADABS.
4) Run SADABS separately for fast and slow scans.
5) Identify overload intensities and edit slow .hkl file.
6) Merge fast and slow scans in XPREP.