Simple disorder using SHELXL: sorbose
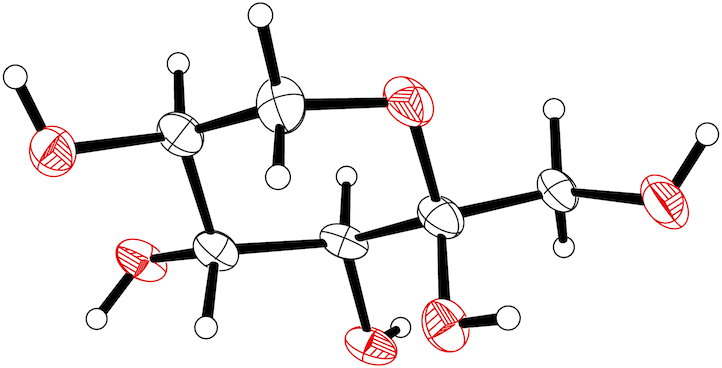
Note: This tutorial uses older programs: shelxs, shelxl97, and shelxtl XP.This tutorial is intended to be an easy introduction to dealing with disorder using SHELXL. In spite of what you may have been told, disorder is not necessarily difficult to deal with, you simply have to know what to do. The structure in question is sorbose, a monosaccharide. Before we get started, it may help to know exactly what we are going to do. The picture below is an ellipsoid plot of sorbose. If you roll your mouse over it, you should see that the CH2OH group on the right hand side shifts to a new orientation. The hydroxymethyl group in sorbose is disordered. In SHELXL, disorder is handled by splitting the atom list into PARTs. 'PART 0' is the non-disordered stuff, 'PART 1' is the major component, and 'PART 2' is the minor component. This tutorial will show you how to do this.