4/4: A SADABS remedy for large b in WGHT
We’ll use a recent gold-containing complex ‘STG-2’ (Gilpatrick et al., 2024) that happened to give particularly weird looking WGHT parameters.
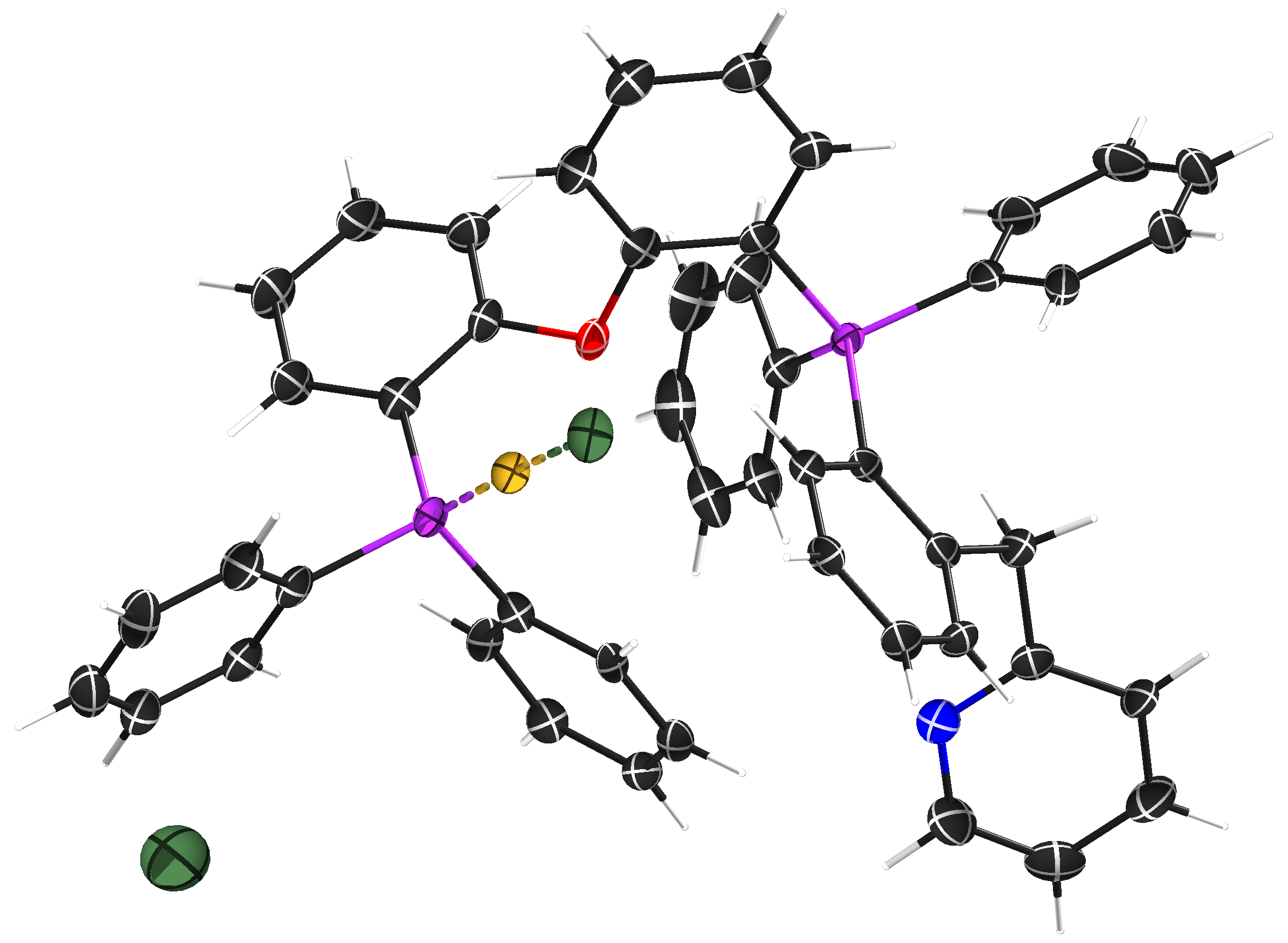
Fig. 2. An ellipsoid plot of gold complex STG-2.
This was a CuKα dataset, which is not ideal for a gold compound (our MoKα optics were broken at the time), but diffraction images all looked fairly good (see Fig. 3) and gave no obvious indication of problems. The published STG-2 made use of SQUEEZE (Spek, 2015) but was otherwise well behaved, so the unusual WGHT parameters were a surprise; related structures were not similarly affected. The *.raw, *.hkl, *.fab, and *.res files are available here.

Fig. 3 Data frames from STG-2 in two different regions of reciprocal space. Nothing seems to be amiss.
Use of default values forced by the APEX gui version of SADABS gave K in the range 0.817-0.984 and a refined g of 0.0447, which in turn gave the following WGHT parameters for a fully refined model:
This default SADABS run gave the Diederichs plot shown in Fig. 4, indicating a limiting I/σ(I) of 22.4. For context, values quoted by Krause et al. (2015) range between 16.4 and 82.5, and the authors suggest a value of ~30 being good for synchrotron data. Thus, the overall data quality for STG-2 seems reasonable for a routine structure.
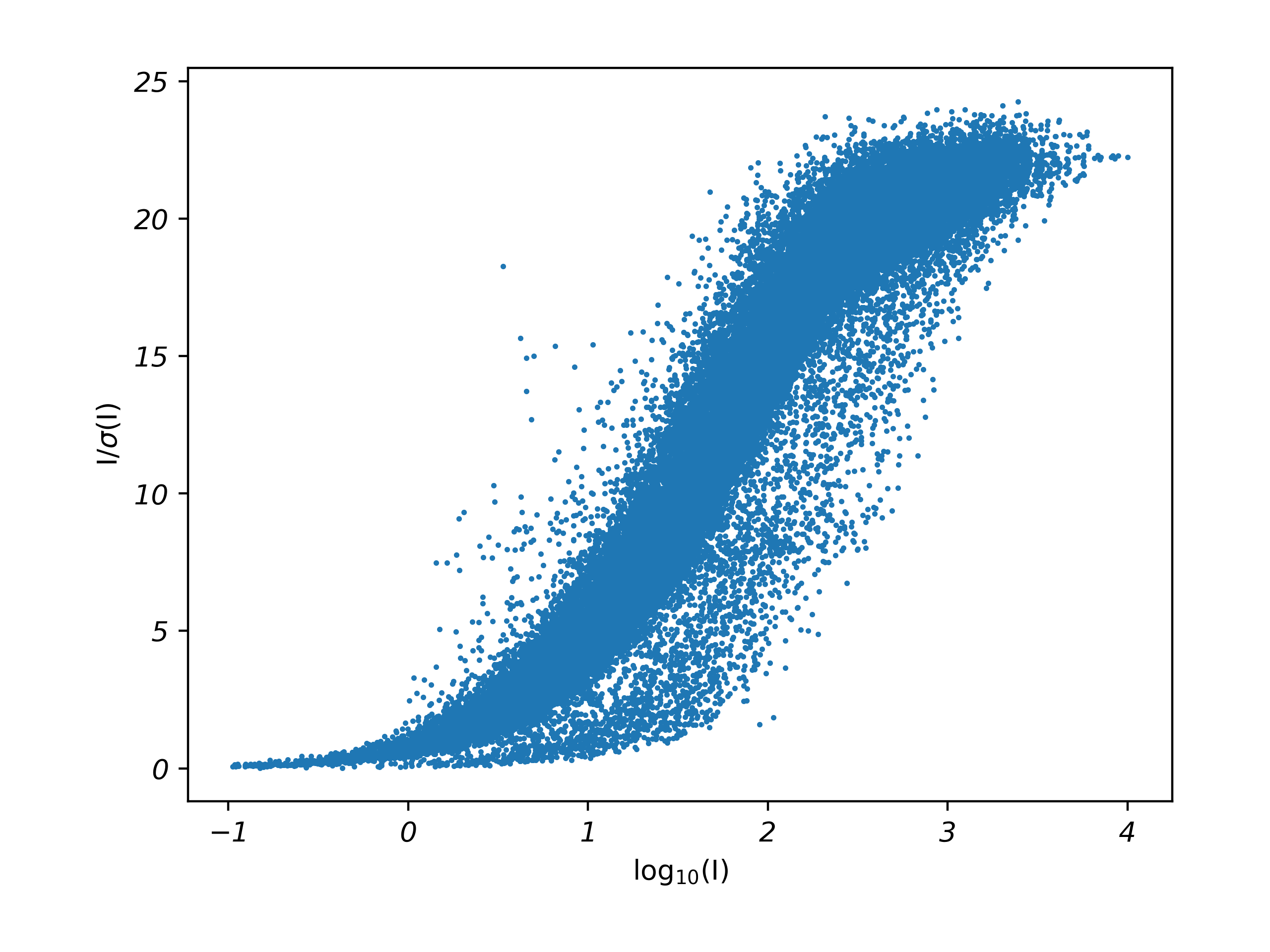
Fig. 4. Diederichs plot for STG-2 from a default SADABS run. Note the abundance of points at the upper right [high I/σ(I)] of the scatterplot.
After a bit of experimentation, setting sm = 3.4 as the 12th parameter on the HKLF command subsequently led to:
The a parameter is still zero, but b takes on a much more normal-looking value. Let’s see if we can achieve something similar using command-line SADABS.
For an initial test we'll try fixed K = 3 and g = 0.0447. It is likely going to take some iteration to optimize so we’ll give each trial a separate filename (e.g., ‘fix-K3-g’), which should help us to keep track of progress. For the first trial, we’ll track the full screen output of the SADABS run as we go and give brief explanations where necessary. The input raw data filename ‘c24009’ is just the sequence number in the UKy X-ray lab logbook assigned to STG-2 when the data were collected. The next few text boxes show the screen output of the command-line SADABS run (highlights indicate changes from defaults).
--------------------------------------------------------------------------
Note that all questions except those asking for a filename etc. may be
answered by "Q" to force SADABS to terminate immediately.
Expert mode (Y or N) [N]: Y
Maximum number of reflections allowed (2000000):
Enter listing filename [sad.abs]: fix-K3-g.abs
Laue group numbers:
[1] -1 [8] -3m (rhombohedral axes)
[2] 2/m (Y unique) [9] -31m (Z unique)
[3] mmm [10] -3m1 (Z unique)
[4] 4/m (Z unique) [11] 6/m (Z unique)
[5] 4/mmm (Z unique) [12] 6/mmm (Z unique)
[6] -3 (rhombohedral axes) [13] m-3
[7] -3 (Z unique) [14] m-3m
[0] to write list of equivalent indices for Laue/point groups to listing file
Enter Laue group number [2]:
Choose ‘expert mode’ to access to all facilities within SADABS and change the name of the log file.
Answering "N" is not recommended unless you have a high redundancy [Y]:
Use a centrosymmetric point group for error model and statistics [Y]:
Equivalent reflections defined by point group 2/m for scaling
Equivalent reflections defined by point group 2/m for error model
Enter name of HKLF 4 format file containing reference or native data to which
the new raw data should be scaled (
for processing SIR data etc.:
Read reflection files written by EVALCCD (with extension .sad specified) or
by SAINT (extension .raw, default if no extension, or .ram for incommensurate
structures). It is important that all files are from the same crystal and
that reflections have been indexed consistently, i.e. that the orientation
matrices are similar (no rows with signs reversed)! Note that XPREP can
reindex a .raw or .sad file transforming the direction cosines. It is also
possible to read in a .mul file from the integration of a twinned crystal
and select just one twin domain for processing using the SAINT partitioning.
Enter filename (/ if no more) [ ]: c24009.raw
Enter filename (/ if no more) [c24010.raw]:
** Cannot open file c24010.raw **
Enter filename (/ if no more) [ ]:
Mean and maximum errors in direction cosine check function = 0.000 0.002
The mean error should not exceed 0.005, and is usually caused by matrix
changes during data processing.
Approximate wavelength, cell and maximum 2-theta (from cosines etc.):
1.54180 27.579 10.746 30.447 90.012 104.374 89.965 149.06
Above, we enter the raw data file that was created by SAINT during integration.
Thresholds should now be specified for excluding reflections from the
parameter refinement; these reflections may still be corrected and included
in the final output .hkl file
55958 Reflections of which 9301 unique; 29.73 data per frame
Redundancy: 1 2 3 4 5 6 7 8 9+
Number of groups: 445 1028 1202 1151 1011 904 641 566 2353
Mean(I/sigma): -inf 0 1 2 3 5 10 15 20 +inf
Number of groups: 68 342 568 585 894 1484 925 575 3860
Enter mean(I/sigma) threshold (must be positive) [1.5]:
Highest resolution for parameter refinement [0.1]:
Factor g for initial weighting scheme w = 1/(sigma^2(I)+(g<I>)^2), where
sigma(I) is estimated by SAINT and <I> is mean intensity [0.04]:
The following restraint esd could be increased for strong absorbers.
Restraint esd for equal consecutive scale factors [0.005]:
Apply angle of incidence correction (Y or N) [N]:
All entries in the above terminal snippet are defaults ...
skip. Radius in pixels for automatic mask generation:
Number of refinement cycles [25]:
Detector (D) or crystal (C) coordinates for spherical harmonics [C]:
Suitable spherical harmonic orders are 4,1 for weak absorption and 8,5 for
strong. Highest even order for spherical harmonics (0,2,4,6 or 8) [6]:
Highest odd order for spherical harmonics (1,3,5 or 7) [3]:
Marquardt damping factor [0.0001]:
Allow for crystal decomposition by B-value refinement [N]:
Apply face-indexed absorption corrections (Y or N) [N]:
51525 Reflections employed for parameter determination
Effective data to parameter ratio = 22.49
wR2(int) = 0.0963 (selected reflections only, before parameter refinement)
Cycle wR2(incid) wR2(diffr) Mean wt.
1 0.0821 0.0652 0.8920
2 0.0612 0.0586 0.9076
----- lines deleted -----
24 0.0545 0.0545 0.9113
25 0.0545 0.0545 0.9113
wR2(int) = 0.0545 (selected reflections only, after parameter refinement)
Repeat parameter refinement (R) or accept (A) [A]:
Again, all defaults (some repetitive lines deleted) ...
Rejected reflections are ignored in the statistics and Postscript plots
(except the detector diagnostics) and in the output .hkl file
Before applying rejections there are:
55958 total and 9301 unique reflections assuming Friedel's law
High resolution limit [0.1]:
|I-<I>|/su ratio for rejection [4.0]:
g-value for use in: su^2 = sigma^2 + (g<I>)^2 (sigma(I) from SAINT).
This is only used for rejections, not for final sigma(I) values [0.0400]:
Reflections rejected for which |I-<I>|/su > 4.00
where: su^2 = sigma(I)^2 + ( 0.04000 <I>)^2 (sigma(I) from SAINT)
55642 total and 9300 unique reflections left after |I-<I>|/su test
Repeat parameter refinement (P), repeat rejections (R) or accept (A) [A]:
su^2 = [K*sigma(I)]^2 + [g<I>]^2 where sigma(I) is from SAINT
K=1, g=0 (0), K=1, refine overall g (1), K=1, refine all g (2),
refine overall K and overall g (3), refine overall K and all g (4),
refine all K and overall g (5), refine all K and all g (6),
refine overall K, input fixed g (7), refine all K, input fixed g (8),
input fixed K, refine overall g (9), input fixed K, refine all g (10),
input fixed K and g (11) [5]: 11
Enter value for K [1]: 3
Enter value for g [0.03]: 0.0447
Here we specify option 11, fix K = 3, and g = 0.0447, which results in the following:
1 55.0 0.0429 0.842 - 0.922 0.894 - 1.114 3.000 0.0447 22.4 4437 2885
2 107.7 0.0361 0.873 - 0.994 0.897 - 1.273 3.000 0.0447 22.4 5898 4044
3 107.7 0.0385 0.806 - 0.958 0.895 - 1.123 3.000 0.0447 22.4 3998 2728
4 -48.1 0.0352 0.879 - 1.062 0.894 - 1.358 3.000 0.0447 22.4 4688 3576
5 107.7 0.0373 0.868 - 0.944 0.894 - 1.181 3.000 0.0447 22.4 4092 2846
6 107.7 0.0403 0.871 - 0.929 0.894 - 1.359 3.000 0.0447 22.4 3890 2690
7 -48.1 0.0379 0.855 - 0.917 0.894 - 1.338 3.000 0.0447 22.4 2470 1873
8 107.7 0.0379 0.831 - 0.959 0.902 - 1.339 3.000 0.0447 22.4 5823 4098
9 107.7 0.0371 0.877 - 0.938 0.894 - 1.341 3.000 0.0447 22.4 3717 2580
10 107.7 0.0384 0.816 - 0.993 0.894 - 1.114 3.000 0.0447 22.4 3675 2497
11 107.7 0.0355 0.837 - 0.953 0.894 - 1.114 3.000 0.0447 22.4 4098 2856
12 -48.1 0.0332 0.849 - 1.051 0.895 - 1.359 3.000 0.0447 22.4 3928 3099
13 107.7 0.0391 0.852 - 0.973 0.897 - 1.329 3.000 0.0447 22.4 4928 3587
I/s(lim) is the limiting value of I/sigma(I) for a reflection of infinite
intensity, and may be used as a criterion for data quality (Diederichs(2010),
Acta Cryst, D66, 733-740. The above statistics are based on all non-rejected
data, ignoring reflections without equivalents when estimating R(int) and K.
Repeat parameter refinement (P), repeat error model (E) or accept (A) [A]:
Notice that the I/s(lim) value is just the reciprocal of g, i.e., 22.4 = 1/0.0447.
Write Postscript diagnostic file (Y or N) [Y]:
Enter name of Postscript file [sad.eps]:
Short (<21 chars) title for Postscript plots [Test]:
Combine detector plots with same 2-theta [Y]:
Spatial display of (I-<I>)/su greater than [3.0] (0 for none):
Repeat (R), write unmerged .hkl (W), merged .hkl (M), .sca (S), XD format (D),
modulated .hk6 (J), testxtl.dat (BioXhit) (T), unmerged with scan and frame
numbers (V) or quit (Q) [W]:
Reflection output file [sad.hkl]: fix-K3-g.hkl
Mu*r of equivalent sphere for additional absorption correction [0.2]: 0.41
Apply lambda/2 correction for a graphite monochromator (2) or lambda/3 for
Ge or Si monochromator (3) or 3-lambda for multilayer optics and Mo, Ag or In
radiation (4) or no correction (0) [0]:
55642 Corrected reflections written to file fix-K3-g.hkl
Estimated minimum and maximum transmission: 0.3664 0.5803
The ratio of these values is more reliable than their absolute values!
Repeat (R), write unmerged .hkl (W), merged .hkl (M), .sca (S), XD format (D),
modulated .hk6 (J), testxtl.dat (BioXhit) (T), unmerged with scan and frame
numbers (V) or quit (Q) [Q]:
Aside: Here we also change the μR of an ‘equivalent sphere’ in order to better approximate the 2θ-dependent component of the absorption surface. The recommendation given by Krause et al. (2015) for a crystal of size 0.3 x 0.2 x 0.1 mm with μ = 10 mm-1 is to use an effective sphere radius of 0.07 mm, which upweights the smallest crystal dimension by ~5.5 times, viz.
0.1x + 0.5 = 0.07(2x + 4)
0.1x - 0.14x = 0.28 - 0.5
x = (0.1 - 0.14) / (0.28 - 0.5)
x = 5.5
Therefore,
This approximation works surprisingly well in most cases. As shown in the above box, for STG-2 the crystal was 0.20 x 0.15 x 0.07 mm and had μ = 8.42 mm-1, giving μR = 0.41. As a result, one might consider using a more aggressive set of spherical harmonics for the absorption correction than the default, but that is fodder for another tutorial.
We now have a newly written dataset ‘fix-K3-g.hkl’ and its attendant log (.abs) file. We can rename a copy of the previously refined model (for STG-2 we’ll also need the .fab file since it used SQUEEZE ) and iterate the refinement until the WGHT parameters converge. This can be done manually, but is easily automated using ShelXle (Hübschle et al., 2011) and presumably other programs (it is also possible to script using bash or python).
Such a test gave the following:
We’re clearly going in the right direction. The a parameter looks reasonable, and b is much reduced, so let’s try K = 4. There’s no need to explicitly present all the SADABS output again, simply repeat the above steps but use K = 4, g = 0.0447, and use the filename ‘fix-K4-g’, which gives:
So, we’re still heading in the right direction. After running a few more trials (again, this could be automated with python /pexpect or bash /expect ) with K increasing by 0.1 each time, the results tabulate, as follows:
refined 0.0000 83.6045
3.0 0.0300 44.0202
4.0 0.0348 11.6088
4.1 0.0354 7.9550
4.2 0.0358 4.4587
4.3 0.0363 0.9453
4.4 0.0355 0.0000
4.5 0.0343 0.0000
It looks as though K of about 4.3 in SADABS produces sensible-looking weights and any further increases in K are overkill. Subsequent refinement will then lead to further optimization of the WGHT parameters, typically lowering b a little more while leaving a largely unchanged. A Diederichs plot for this treatment is shown in Fig. 5.
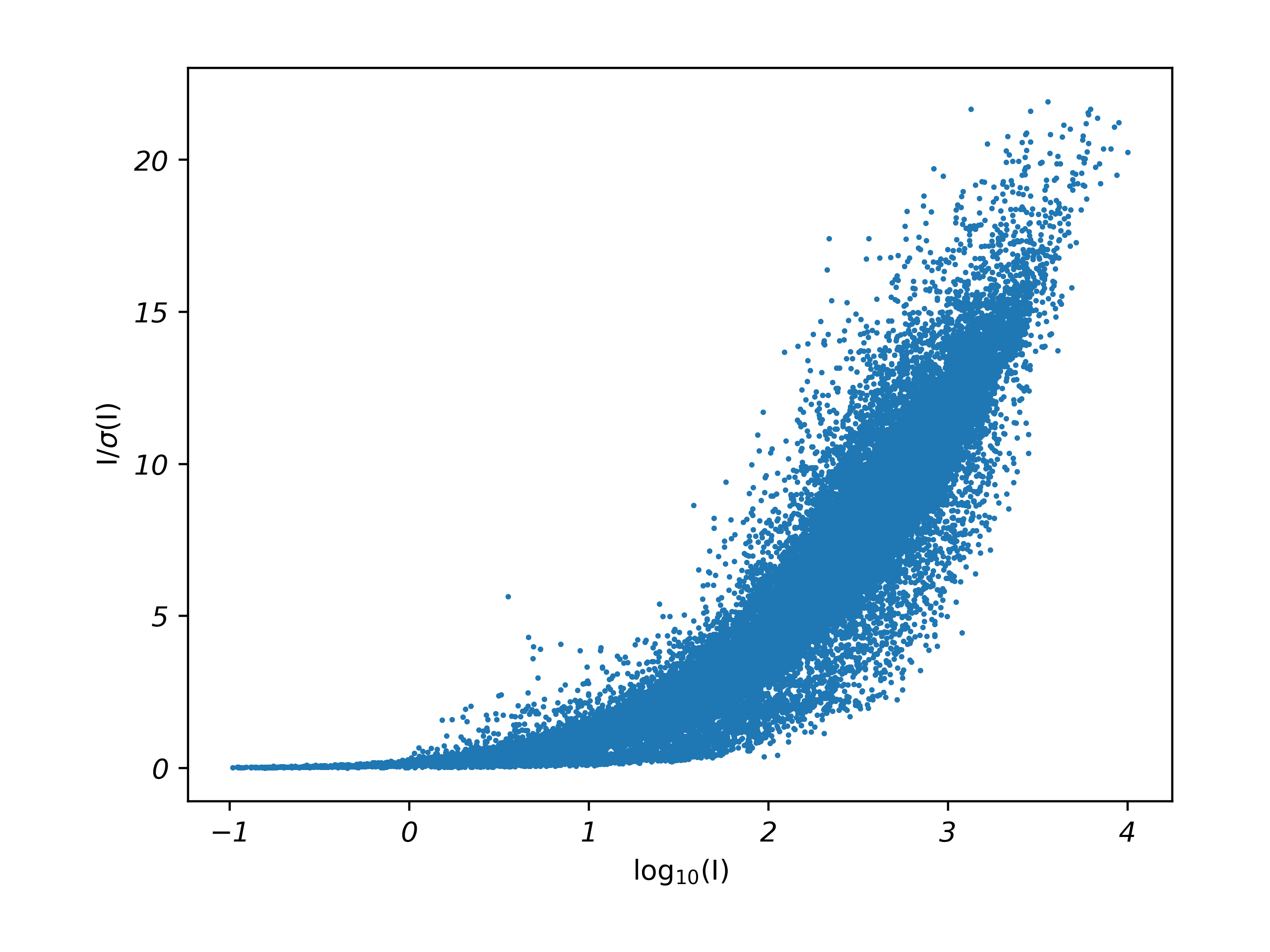
Fig. 5. Diederichs plot for STG-2 from a SADABS run using K = 4.3 and g = 0.0447.
Note that the height of the scatter plot in Fig. 5 matches the default run in Fig. 4, but many of the points are shifted to lower I/σ(I), leaving the plot a little sparse in the upper right. That’s to be expected, as the σ(Fo2) had been previously diagnosed as underestimated.
The high b problem is thus 'fixed' for STG-2, while preserving most other details, including the 'flatness' of the analysis of variance (see *.lst file) and the overall I/σ(limit). This was an extreme case; few large-b WGHT problems will require such a big K value in SADABS. Something like a K of 1.1 — 1.3 is more typical.
Aside: It's important to note, however, that little of this makes much difference to the resulting structure model. Does any of it matter ? I don't know. My own view is that it might be preferable to leave it all as is so that everyone has the same information, not hidden in a SADABS log file. I've always been a bit leery of the 'forcing the goof = 1' notion. If there's a bona fide statistical basis for that, it's alien to me, and (apparently) much of the statistics community. The goodness-of-fit should be 1 for distributions drawn from the same population. Observed and calculated F2 don't fulfill that criterion, especially if the spherical-scattering-factor approximation is used (that's most structures). It's silly to think that they should.
The last bit of that comes from my notes as a grad. student in 1992 when we first got a beta version of SHELXL-92 at UC Davis. They are basically a paraphrase of Håkon Hope's comments in a group meeting. (Message to students: keep copies of your notes.)