4/6: Find true space group symmetry using Platon
To check for missed symmetry, we'll use
ADDSYM in
Platon. For this,
Platon
needs a
cif and a 'LIST 4' format
fcf file, which are written by
SHELXL if
both 'ACTA' and 'LIST 4' are present in the
ins file. These files:
monI.cif
monI.fcf
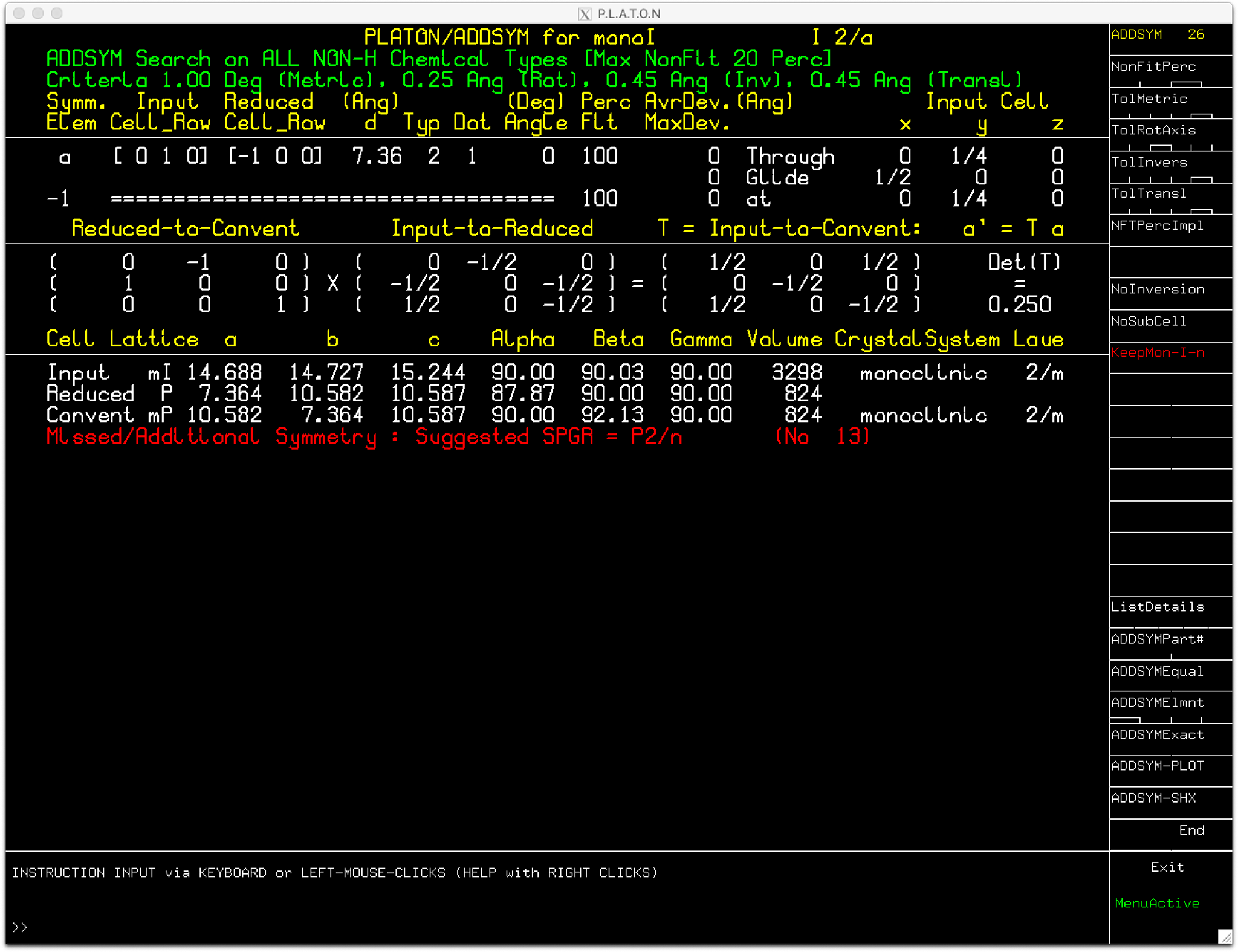
On reading the
cif into
Platon and clicking 'ADDSYM' (top of the SYMMETRY
column), we get the following:
ADDSYM suggests a primitive monoclinic cell with a quarter the volume of the
I-centered
cell. [Note: At the time of writing (Nov-Dec 2020), the latest available version(s) of Platon might
require you to click on one of the options on the right hand column (e.g. NonFitPerc) before it makes
the transformation to monoclinic-
P ].
It also (on the lower right) gives the option to write out a new
SHELXL res file with
the structure transformed for
P2/n. Go ahead and create the file as we'll use it later - it
will have '_pl.res' appended to the file name stem. A copy is available here:
monI_pl.res
ADDSYM also supplies a matrix to transform from the
I-centered cell to the
P2/n cell:
0.5
0
0.5
0
-0.5
0
0.5
0
-0.5
The above transformation generates many non-integer reflection indices. Normally, that is not a
problem as they'd be simply ignored or deleted; non-integer indices do not represent actual Bragg
peaks. The twinning, however, causes such 'impossible' reflections to coincide with Bragg
maxima from other twin domains, so we cannot not simply discard them.
The relationship between the
I-centered tetragonal pseudo-cell and the true monoclinic-
P
cell of each individual is shown below:
There is no way to take account of the four individuals for this structure using the TWIN command
in
SHELXL while maintaining the correct space group symmetry, even with an unconventional
setting. The way forward is to make a data file in 'HKLF 5' format that preserves all the information.
At the time of writing, I'm not aware of a way to do this in
Platon without first transforming
to
P2/n. Unfortunately, for the reason mentioned above, that would eliminate about half the
observed intensities. In section 5, 'Generate 'HKLF 5' format dataset', we'll manually prepare an
'HKLF 5' format file. It's a bit tedious, but not difficult.