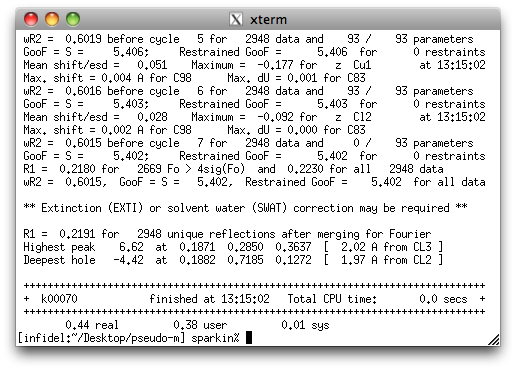
As you can see, it does not look very good. Hopefully, adding TWIN and BASF
instructions will help.
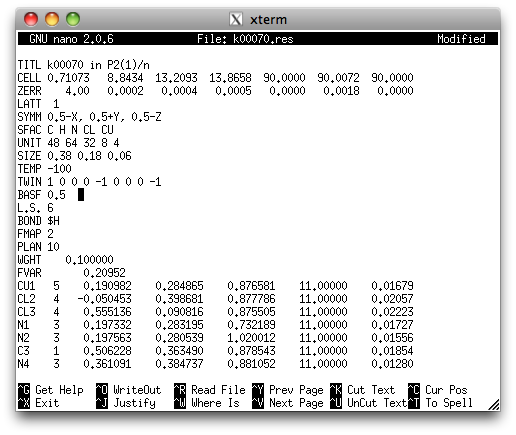
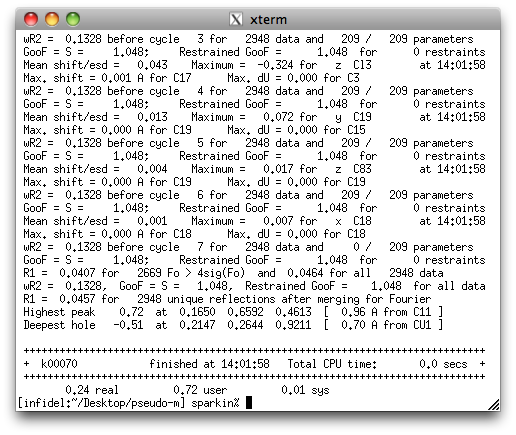
A few cycles of refinement with SHELXL reveal the following:
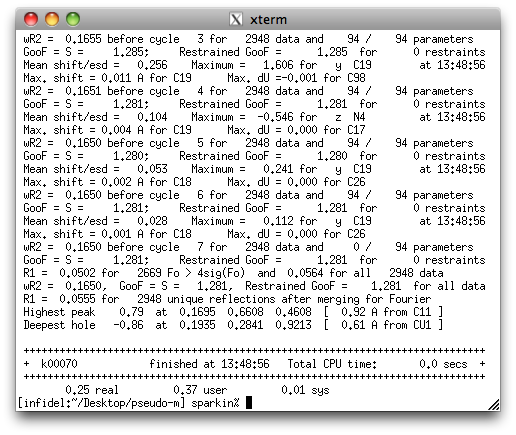
That's a whole lot better. The R-values and GooF are much improved, and the
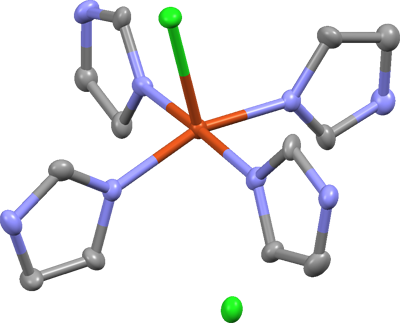
difference map is considerably flatter. Here's what it looks like in Mercury:
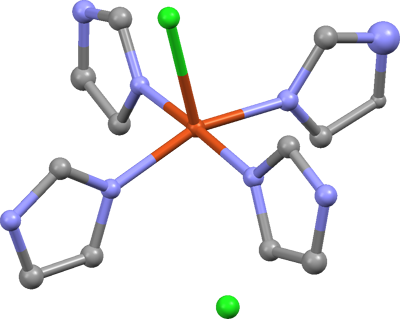
The model is dramatically better, but a nitrogen and carbon atom on the upper right
ring seem odd. The N looks too big, and the C looks too small. This
illustrates the effect of wrongly assigned atom types. In this case, the N
has been smeared out by the least-squares in order to fit the experimental electron
density of carbon. Likewise, the C was 'sharpened' to better fit the
scattering power of nitrogen. Manual editing of the next '.ins' file is sufficient
to switch their atom types. In SHELXL we only need to change the
scattering factor number in the '.ins' file, but for aesthetics we'll also change
the atom labels). While we're at it, we'll also make the model anisotropic
(ANIS).
Yet more improvement. It is now time to re-check atom assignments prior to applying
a sensible numbering scheme and adding hydrogen atoms to complete the structure.
We'll renumber the structure using XP, as usual ...
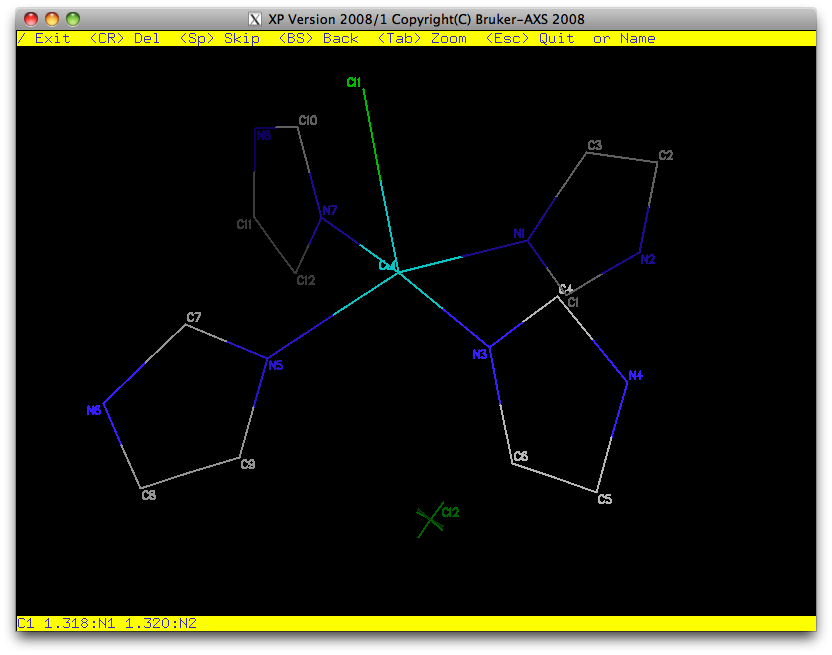
As with any structure, it helps to use a sensible numbering scheme, and to sort the
atoms in a logical order (SORT in XP). We'll save this model, run a
round of refinement and see if hydrogen atoms show in the difference map. This is
the result:
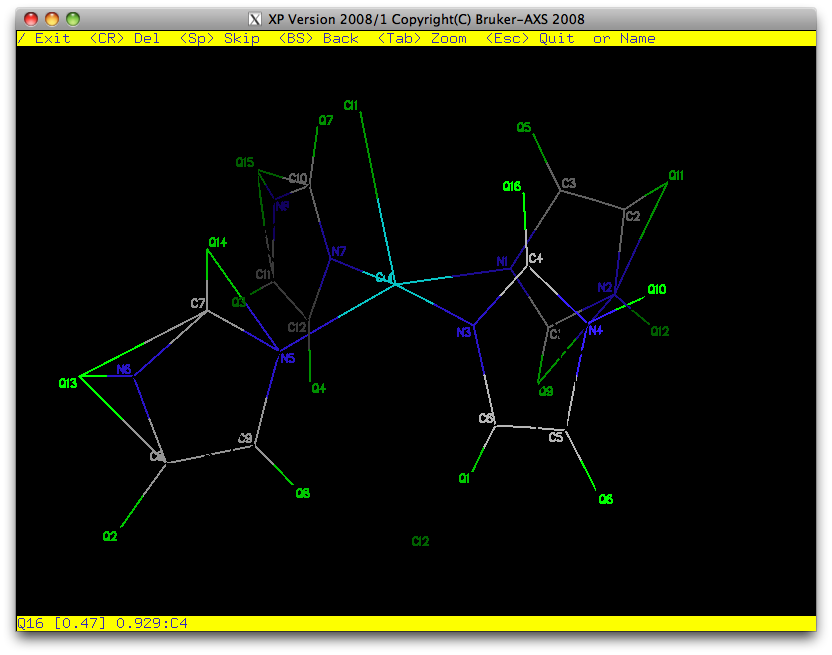
For sure we can add hydrogen atoms to the carbons ...
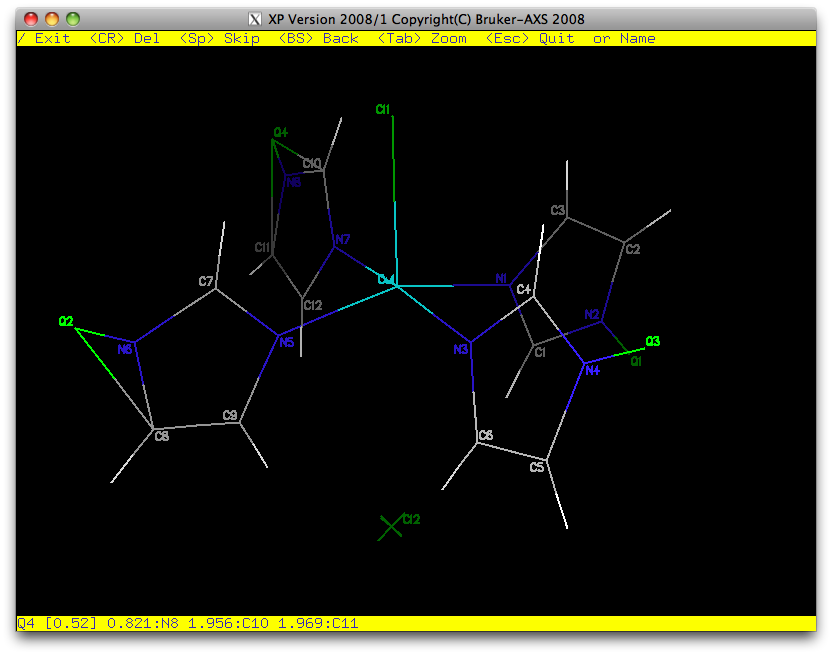
After another round of refinement, nitrogen atoms N2, N4, N6, N8 also clearly have
hydrogen attached. We'll add those to the '.res' file, refine, modify the weighting
scheme (WGHT instruction), check to see that the moieties are placed well
in the unit cell and test for extinction effects (EXTI).
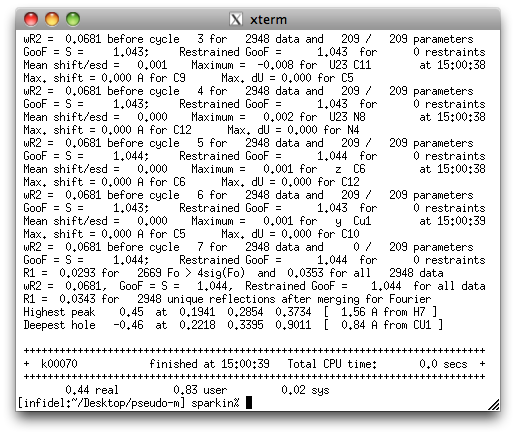
A final round of refinement shows that in spite of the twinning, the structure is
rather high quality. The '.res' file is available here.
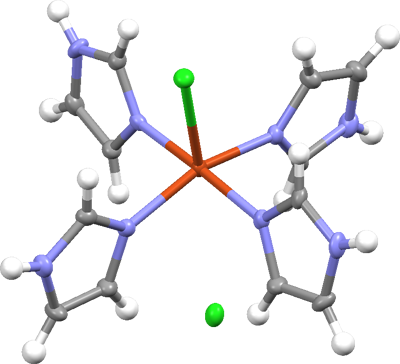
Conclusion: The final refined structure model is good, with no unusual features.
1) Assign a space group using XPREP.
2) Direct methods structure solution.
3) Decipher the twin law.
4) Twin refinement with SHELXL.
2) Direct methods structure solution.
3) Decipher the twin law.
4) Twin refinement with SHELXL.
Return to the first page of this tutorial.