4) Elementary model editing in maXus
You are now ready to edit the model. In this laboratory it is common to do atom assignment from within maXus and then to run a few cycles of refinement before proceeding to complete the structure with the SHELX and SHELXTL (XP) programs.
To edit the results of your structure solution, click "Model" and choose "display" from the sub-menu.
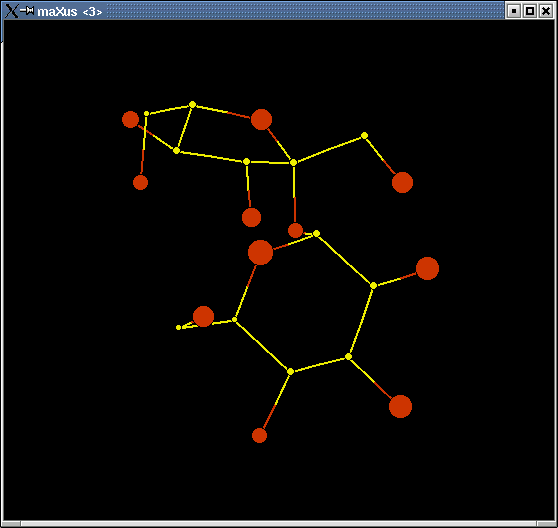
Note that you could have clicked "display expanded" - that would have given you symmetry-related "atoms" as well. Often it helps to see these in addition to just the asymmetric unit - it depends on the structure. If you do "display expanded", the square-shaped "atoms" are the symmetry generated ones. In any event, you will see something like this:
 |
 |
 |
|
This is actually three separate windows. Often the structure will not look nearly so "clean" as this one!
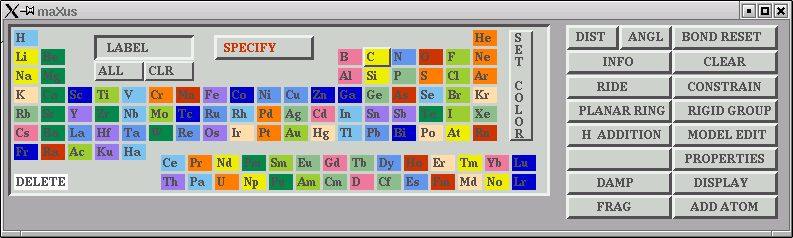
You can spin the molecule around using your mouse to drag it. To get rid of false "atoms", click on the "DELETE" button on the lower left and then click the "atoms" you want to delete. To rename an element type, click on its symbol on the periodic table and then click on the atom site on the model.
Other facilities are available to you here also, such as checking bond lengths and angles etc. Generally speaking, however, in this laboratory we only tend to use the model editing features, but feel free to experiment (but don't expect much help other than from the manual).
When you are happy (or reasonably happy) with your molecular fragment(s), click the "SAVE & EXIT" button and you will be safely transported back to the maXus main menu:

Here you can safely skip "SFac" and go straight to "Refine". Our aim now is simply to do a few cycles of least-squares refinement and prepare a model file that is in SHELX format (these are the *.ins and *.res file that you have already seen in the lab.)
Under the "Refine" menu item you should choose the SHELXL option and go for the default, which will give you something like this:
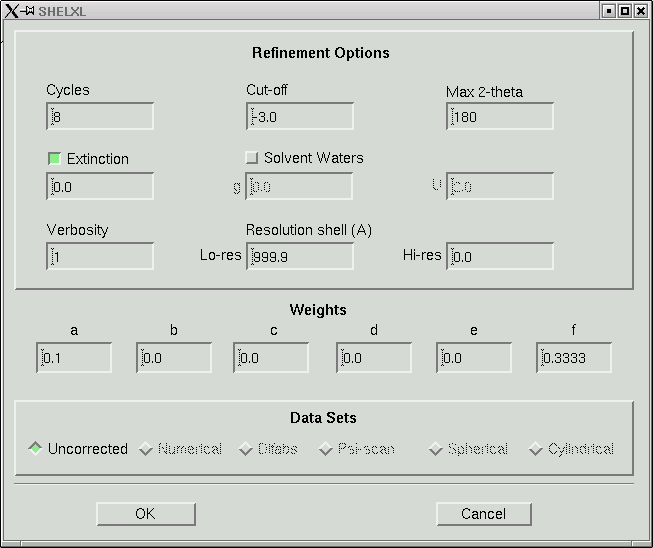
In the old days when computers were slow, 8 cycles of least-squares would take a long time for any but the smallest structures. Even though computers are fast now, there is still no point in wasting computer time. Change the number of cycles to 4, like this:
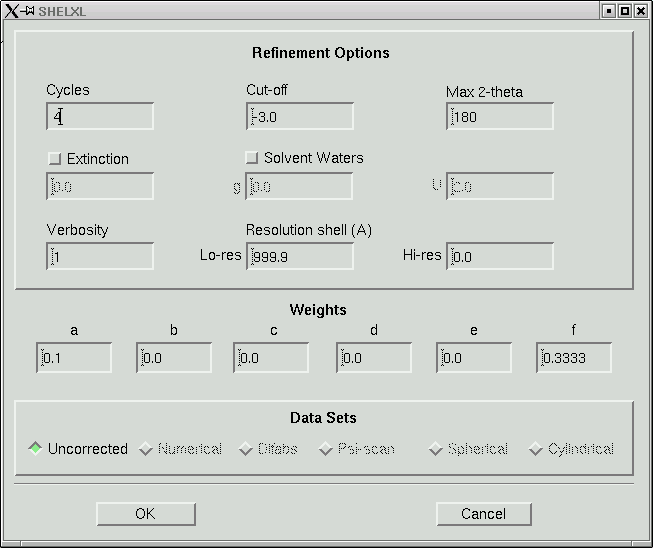
Then click "OK". The maXus program then spawns a SHELXL job to refine your model. It outputs the usual SHELXL statistics into a window like this:

Things to look for in this output include:
1) Values for wR2 that get smaller down the list. They may also stabilize (as above), which means the refinement has converged.
2) The GooF should get closer to 1.000, but it will not likely get very close to 1.000 until later in the refinement when you manipulate the weighting scheme.
3) The "Mean shift/esd" should get smaller down the list.
4) Max. shifts should also get smaller down the list.
5) The largest difference map features (peaks and holes) give useful information on what to for subsequent refinement rounds.
... and finally it finishes with the following message:
Click on "OK".
You now have a file entitled "shelxl.res" within a directory called "maxus" inside the directory (e.g. k06123 or whatever) that you created way back at the beginning of this structure determination.
It is probably a good idea now to exit from the maXus programs and go back to the terminal window to do a bit of sensible book-keeping prior to completing the refinement with SHELXL and XP (the SHELXTL molecular editor). The steps involved are:
1) Exit from maXus.
2) Go back to the (or open a fresh) terminal window and make sure you are in the directory that contains all your data collection (*.kcd) files.
3) Run "cifin" and create a file named (say) k06123.hkl - it should be obvious where to type what into the dialogue box.
4) Create a separate directory in which you will keep all your subsequent refinement files, perhaps like this:
mkdir refine
5) Copy the *.hkl file that you just made with "cifin" into that directory like this:
cp k06123.hkl refine/.
6) Copy the shelxl.res file from the maxus directory into your refine directory and change its name as you do so:
cp maxus/shelxl.res refine/k06123.res
7) Copy the nreport.html file into your refinement directory:
cp nreport.html refine/.
8) You really don't need all those large *.kcd files anymore but they do need to be archived onto CD. In this lab we always archive all data. Compress them like this:
bzip2 *.kcd &
This takes a while because these files are large and there are usually a lot of them. You do not need to wait, however, go ahead and change your directory ...
cd refine
... and proceed to complete your refinement with "xp" and "shelxl".
Go back to maXus structure solution guide section 3
Go back to maXus structure solution guide main menu
Return to the main Tutorials page or to the main X-Ray Lab page