13) Prepare a set of data collection scans.
Back to the SuperGUI menu - notice that the "Guess diffraction limit" button is now green. Click on "Collect images"
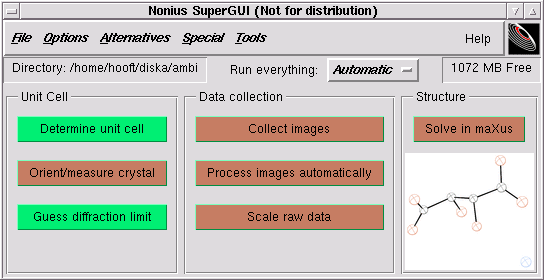
In the following window, click on "Make Scan-set" ...
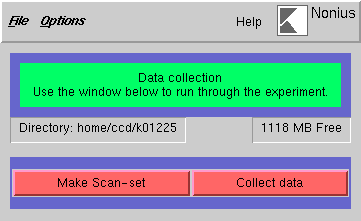
... to bring up this window:
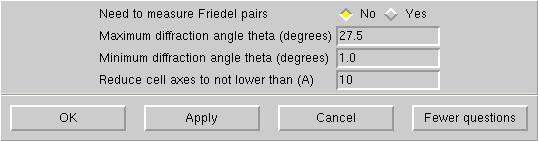
If your crystal is small or diffracts poorly, you may want to reduce the maximum theta to 25° or even 22.5°, but no lower than that. Click "OK" to get this:
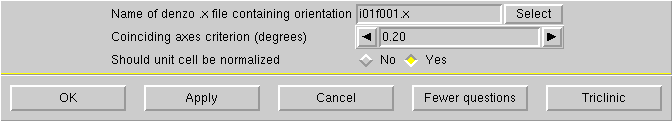
Unless you have some good reason to do otherwise, click "OK" and you will be prompted to make a connection to the diffractometer again:
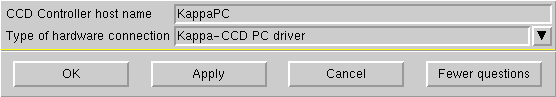
Click "OK" to begin the set-up of data collection scans:

When you click on "Strategy" the following window appears:
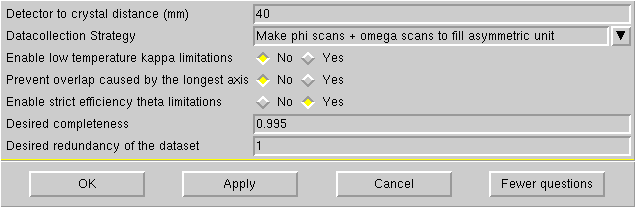
If the longest cell axis < 20Å, then a distance of 40 mm will almost certainly be fine.
For larger cells, increase the distance so that in millimetres it is about twice the longest cell edge in Å (i.e. for a 30Å cell edge, set the distance to 60 mm).
Do NOT set the distance less than 40 mm - you will regret it if you do.
The default for our kappaCCD is different from the above example. We always make ω scans at fixed χ = +/-55° and if needed, a fast φ scan (at χ = -55°). In fact the defaults that are set on our machine are almost always ok, and if they are not ok for your crystal (other than the need to add a fast φ scan) you will probably need professional help.
Click "OK" and the program calculates a bunch of scans that fit the criteria you just specified. It then lets you examine them:
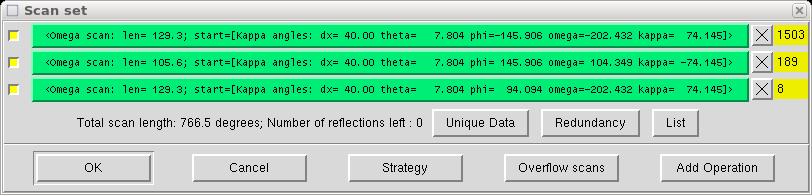
You can see how many new and unique reflections are obtained in each scan and check redundancy, which is a measure of how many times some reflection (hkl) or an equivalent is measured.
Lower symmetry requires more scans to cover the unique part of reciprocal space. Nine scans are typical for triclinic, five or six for monoclinic and three for orthorhombic crystals etc.
Now we get to add the special fast φ scan to take care of overloads. In the above "Scan set" window click on "Add Operation", which brings up the following:
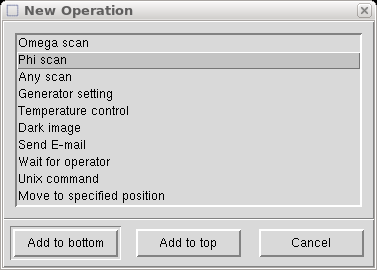
Select "Phi scan" and click on "Add to top", and you get the following:
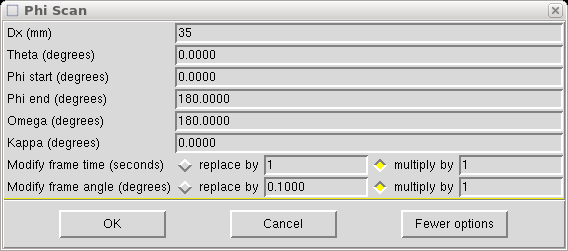
Which you need to modify as follows:
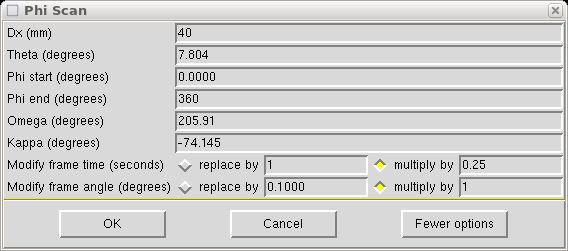
The Dx value is set to 40mm in this example. If your main scans are set at some other Dx value (e.g. greater than 40mm if you have a cell edge >20Å) then set Dx to that value. Set theta to the same as the main scans, which in this case is 7.804°. Set φ to do a full 360° rotation. The ω and κ values are set to position the φ axis (i.e. the mounting pin) perpendicular to the x-ray beam, but angled at χ = -55°. We do this to ensure that the fast φ scan keeps the φ axis well away from collinearity with the cold stream of the low-temperature machine. There is a bit of leeway in the angle, but the above values work well so don't change them without good reason. Remember: with a kappa-axis goniometer, ω and κ are not orthogonal, so if you change one you will have to change the other to keep the φ axis perpendicular to the x-ray beam [(-55° χ, 180° ω) = (-74.145° κ, 205.91° ω)].
You will also need to change the scan time per frame - either to an absolute value or as a fraction of the main scans frame duration. Here we make it a quarter. Something between 0.1 and 0.25 is usually ok. The point is to collect the fast frames so that the most intense reflections do not overload the detector.
When you click the "Ok" button, your new scan appears in the list, like this:
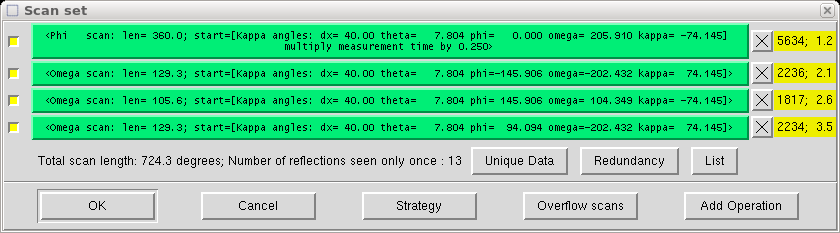
Click on "OK" when you are satisfied.
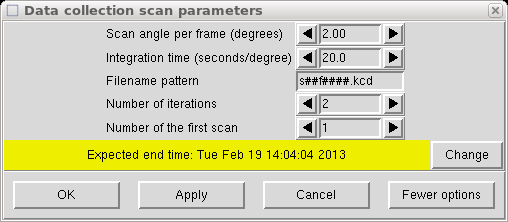
Now we get to choose the width and duration of each frame. Scans of 2° are ok most of the time but with long cell axes and mosaicity > 1°, it is usually best to reduce it to around 1°. If the program suggests less than 1°, seek professional help. There is still some healthy debate about whether wide scans or narrows scans are best. Most (but not all) of the arguments in favour of narrow scans are historical in nature, and are irrelevant for most crystals. There are pros and cons to both approaches, but narrow scans will always use up a lot more disk space.
Twenty seconds is usually ok but weak/small crystals may need more. Greater than 120 seconds is usually a bad idea. The number of iterations should be 2 to help eliminate zingers.
The final stage is to start data collection ...
Return to the main Tutorials page or to the main X-Ray Lab page