3/6: Solve structure in I 2/a using XPREP
SHELXD often gives interpretable models for twinned structures, but performance is usually
better if some (at least partial) information about twinning is supplied. At this stage, the actual
nature of any twinning would not normally be known, but a reasonable guess might be rotation about
the pseudo-tetragonal
c-axis. The following matrix describes a 90° anticlockwise
rotation about
c :
Repeated application generates four plausible twin components.
SHELXD is able to use a TWIN
matrix but the current version (
SHELXD-2013/2 ) is restricted to two components set
via BASF (default 0.5). Still, partial information is better than none, so we'll include a second
domain. In the following, a 'TWIN' command corresponding to a two-fold (i.e., 180°, or 90° applied
twice) about
c has been added (feel free to experiment with 90° or 270°).
The structure is quite small, so even with 'NTRY 1000' it runs quickly. The above SHELXD
ins file and the dataset transformed for the I-centered setting are available here:
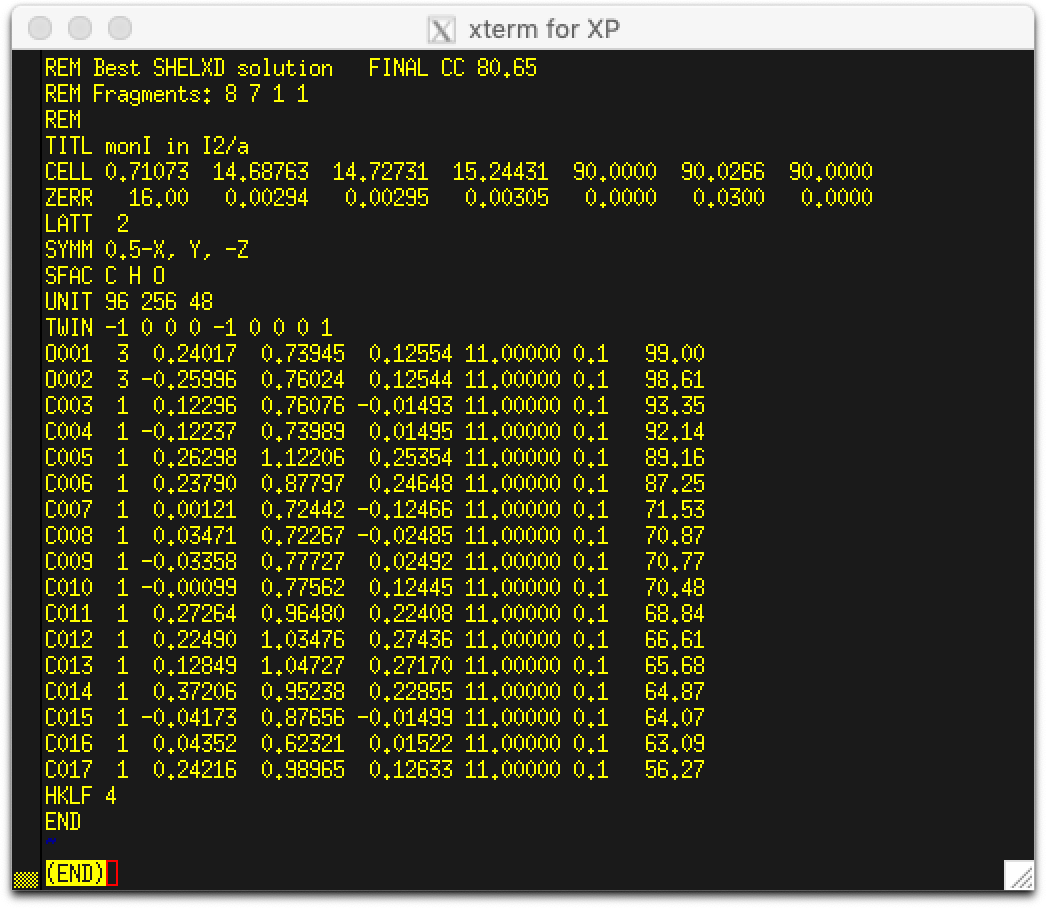
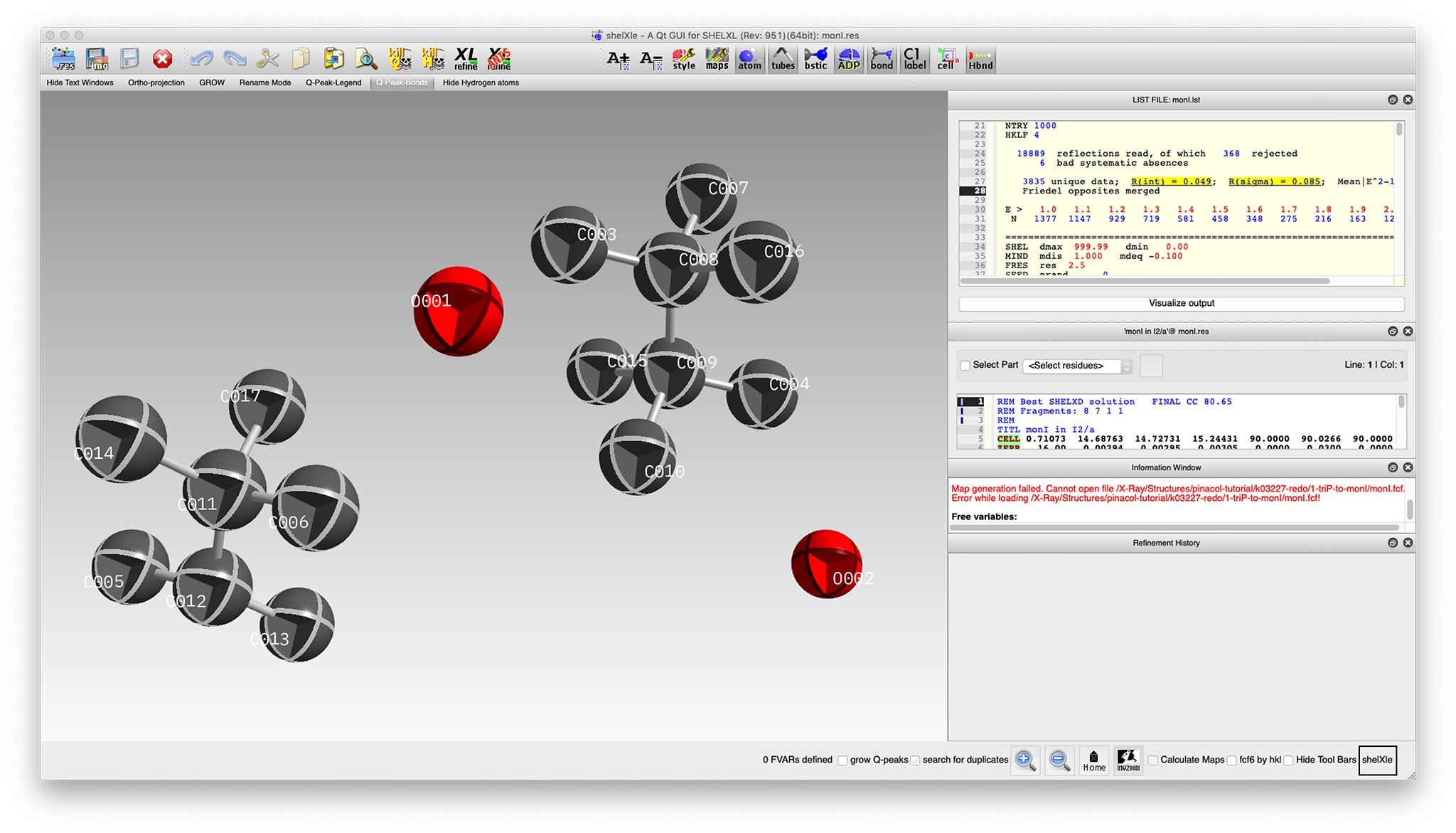
On completion of the SHELXD run, a res file something like the following is written:
When viewed in ShelXle, it looks like this:
Most of the atoms for a pair of pinacol molecules are present (albeit with mis-assigned oxygens)
along with two water oxygens. For this structure,
SHELXD sometimes gives a model with two
pinacol fragments (as shown), or it might give four half molecules, each situated on inversion
centres. At this stage it does not matter, as neither are crystallographically 'correct'. Either
'solution' can be 'fixed' in much the same way.
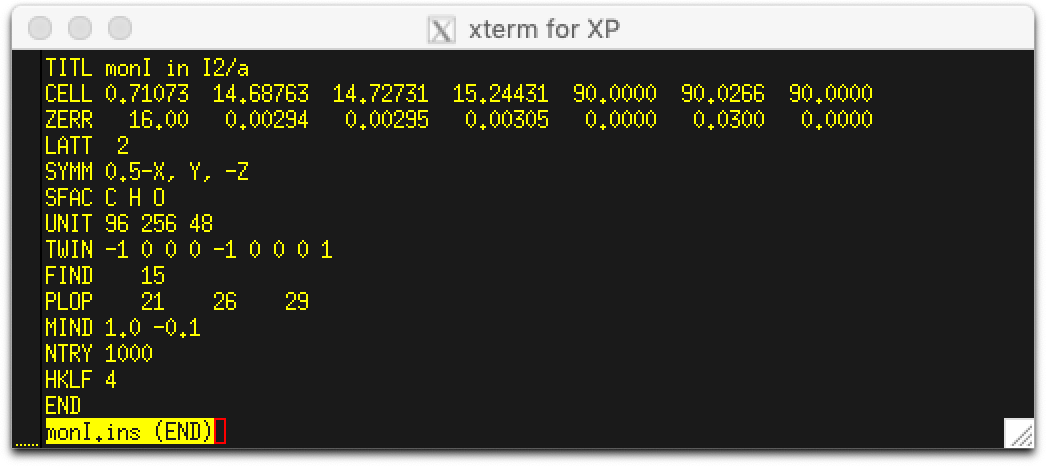
The
res file written by
SHELXD needs editing before refinement. At the very least it
needs L.S., FMAP, PLAN, FVAR, and BASF instructions. Since the twinning is not yet treated properly,
we can expect the refinement to need extra help. Thus, a severe SIMU restraint, say 'SIMU 0.005' is
advisable (at least temporarily), something like this file:
monI.res
A few cycles of isotropic refinement using the above
res file followed by model building to
fix the pinacol oxygen atoms gives something like this:
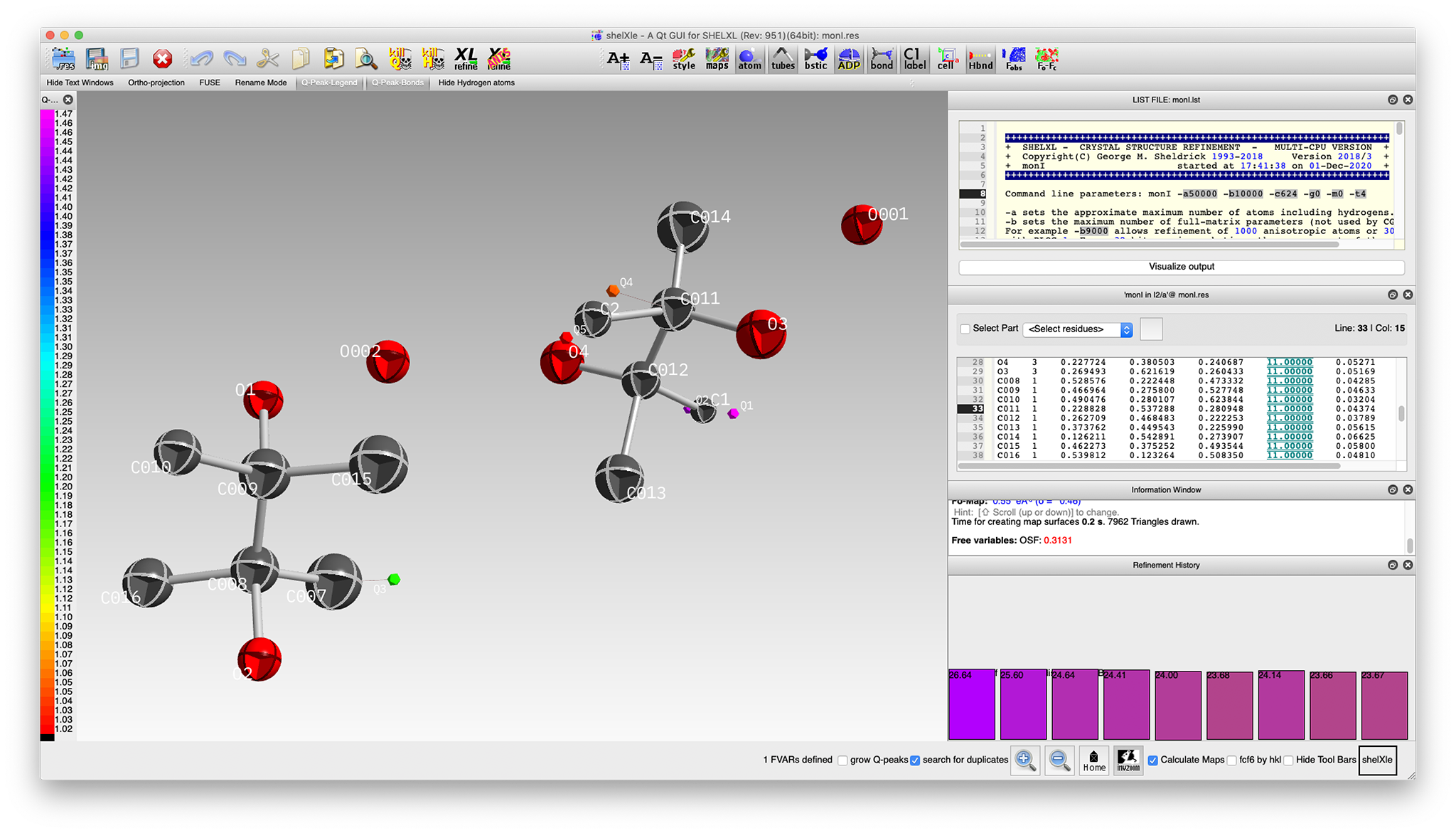
That looks a whole lot better, but refinement statistics are awful! In particular,
the R-value is stuck at about 24%. A major reason is that the model still has incomplete
treatment of the twinning. We can easily test our suspected four-fold twinning by modifying the
TWIN and BASF instructions to cycle through each 90° step of the four-fold, like this:
TWIN 0 -1 0 1 0 0 0 0 1 4
BASF 0.25 0.25 0.25
Note here the change to the TWIN operation to describe a 90° rotation. Also, those three BASF
parameters are just initial guesses and will refine. After a few cycles, the model is dramatically
improved. Even anisotropic refinement (with restraints) is stable, as is addition of riding methyl
hydrogens. Consequently, the R-value plummets to ~6.5%
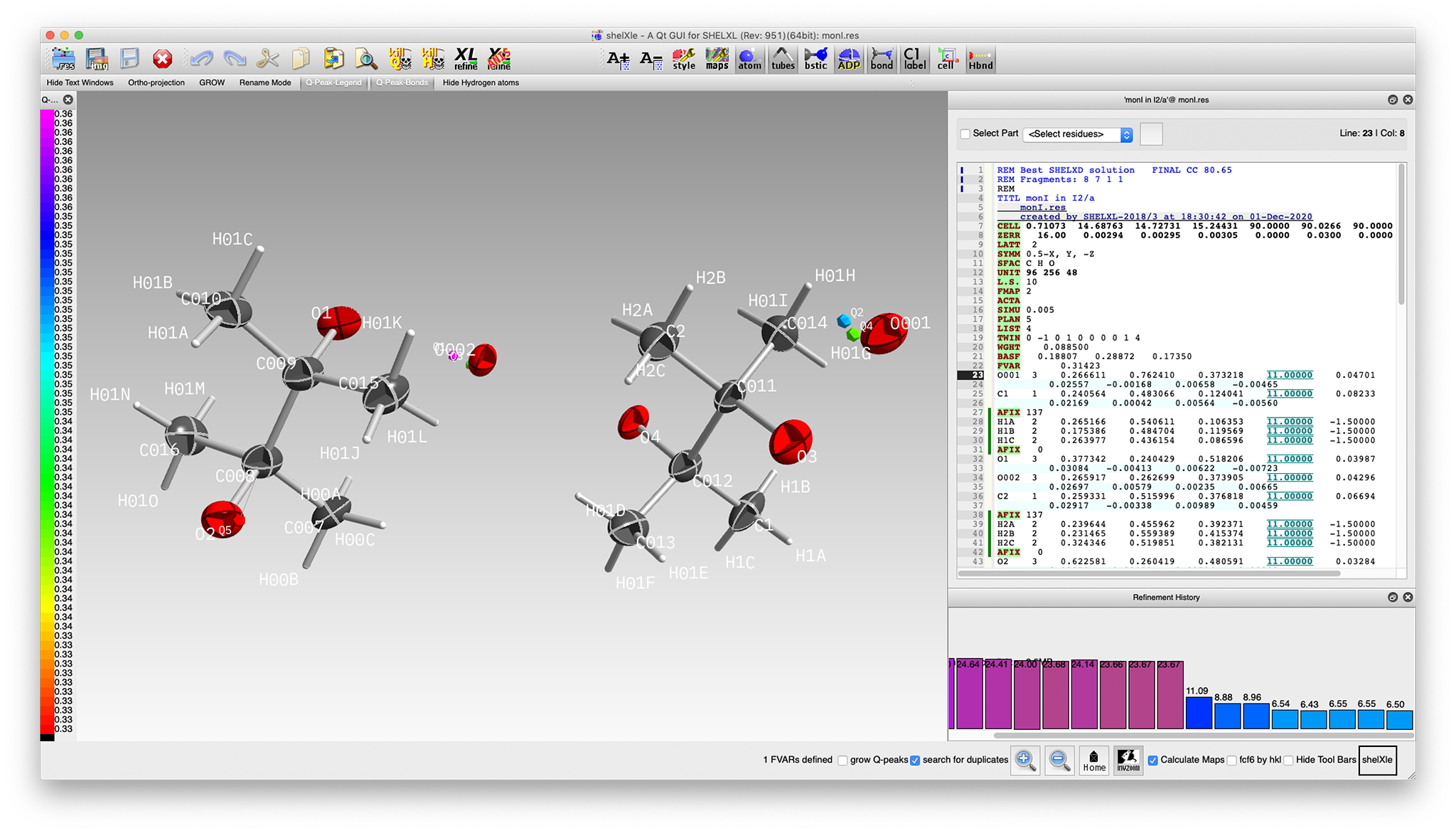
In spite of how good this looks, the model still has problems. A careful inspection reveals hefty
correlation between atoms related through the centre of each pinacol. In this I 2/a,
model, those are pseudo-inversions. With hindsight, in the true space group (P 2/n),
they're real crystallographic inversion centres, so further work is necessary. In the next section
'Find true symmetry in Platon', we'll use ADDSYM within Platon to check for
missed symmetry.