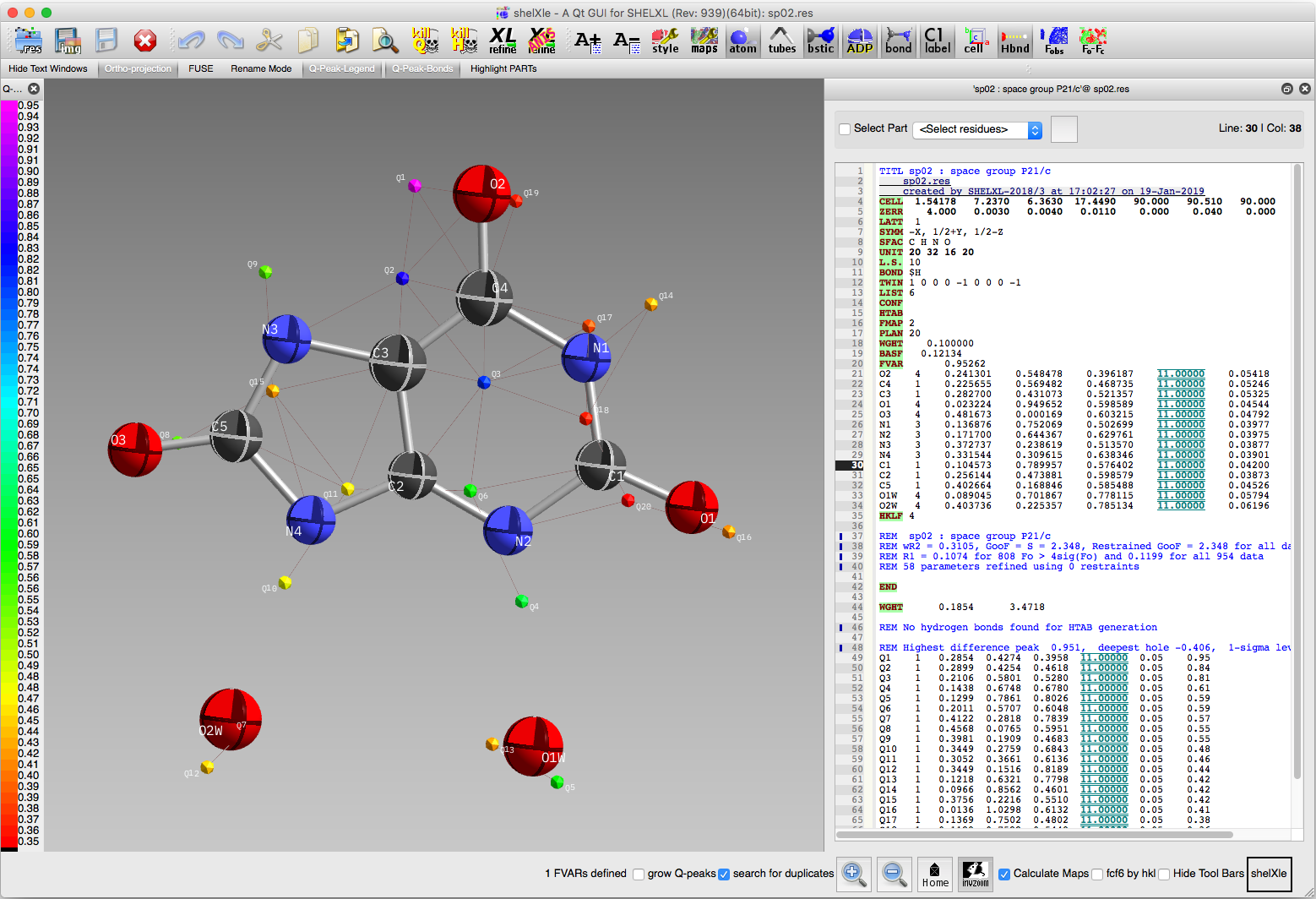
In the above image, the three largest difference map peaks provide the key
to how this molecule is disordered. Note how peaks Q1 and Q2 are positioned
just to the left of atoms O2 and C4, while peak Q3 is just to the right of C3.
If we were to flip the whole molecule by 180° about a vertical axis in the
plane of the image, the new positions for O2, C4, and C3 would coincide with
difference map peaks Q1, Q2, and Q3. C2 and Q6 also appear to be related by
the same operation. From the looks of it, such a 180° flip for the water
molecules would superimpose O1w on O2w and vice versa, which means we don't
need to model disorder for the water molecules. We therefore proceed by setting
up for the next round of refinement with a fragment (FRAG) consisting of
a whole uric acid molecule, and instructions to fit O2, C4, and C3 of the minor
fragment to the positions of peaks Q1, Q2, and Q3. To make it easier to follow,
we'll rearrange the atoms a little to place O2, C4, C3 at the top of the list.
The edited file looks like this:
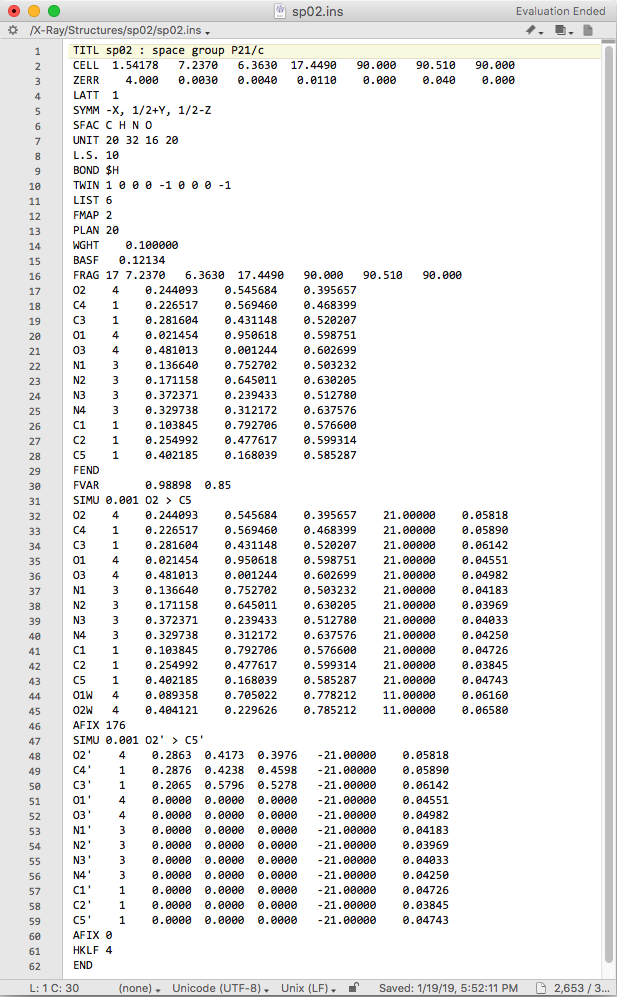
Ok, what to make of that! We'll break it down, step by step:
1) The FRAG line has code 17 (see SHELXL manual) and is followed by the unit cell dimensions. Here, the unit cell is needed because the fragment is given in fractional coordinates. If the fragment were defined by Cartesian coordinates, the unit cell would not be needed on the FRAG line.
2) Coordinates of the fragment atoms, in the same order as the main component. This is merely a copy of the uric acid atom lines, but with occupancies and displacement parameters removed (they're removed only for the sake of clarity, as SHELXL would ignore them if we'd left them in - see the SHELXL manual!).
3) FEND marks the end of the fragment.
4) A second number has been added to the FVAR instruction. It is used to define the occupancy factor for the major component, and it be will refined. The minor component is set so that the sum of occupancies equals 1.0.
5) As a precaution, strong SIMU restraints are applied to displacement parameters of the disorder components. If all goes well, we'll relax this later on.
6) The next 14 lines are the atoms of uric acid and water molecules. Note that the occupancies for uric acid are set to the second FVAR parameter, while the waters remain fixed at full occupancy.
7) The AFIX 176 ... AFIX 0 section defines which atoms are to be used in the rigid-body fragment fitting and which are just along for the ride. In this case, the first three atoms (O2', C4', C3') have been assigned the coordinates of Q1, Q2, Q3 difference map peaks. NOTE: You need to assign at least three atoms for this to work - bonus points if you can explain why! All other uric acid atoms are given zero coordinates. SHELXL interprets this as "keep the geometry of the fragment the same, but superimpose it on the major component by fitting the (three) specified atoms". That's the plan at least! Let's run a round of refinement and see what happens. The view in ShelXle should look like this:
1) The FRAG line has code 17 (see SHELXL manual) and is followed by the unit cell dimensions. Here, the unit cell is needed because the fragment is given in fractional coordinates. If the fragment were defined by Cartesian coordinates, the unit cell would not be needed on the FRAG line.
2) Coordinates of the fragment atoms, in the same order as the main component. This is merely a copy of the uric acid atom lines, but with occupancies and displacement parameters removed (they're removed only for the sake of clarity, as SHELXL would ignore them if we'd left them in - see the SHELXL manual!).
3) FEND marks the end of the fragment.
4) A second number has been added to the FVAR instruction. It is used to define the occupancy factor for the major component, and it be will refined. The minor component is set so that the sum of occupancies equals 1.0.
5) As a precaution, strong SIMU restraints are applied to displacement parameters of the disorder components. If all goes well, we'll relax this later on.
6) The next 14 lines are the atoms of uric acid and water molecules. Note that the occupancies for uric acid are set to the second FVAR parameter, while the waters remain fixed at full occupancy.
7) The AFIX 176 ... AFIX 0 section defines which atoms are to be used in the rigid-body fragment fitting and which are just along for the ride. In this case, the first three atoms (O2', C4', C3') have been assigned the coordinates of Q1, Q2, Q3 difference map peaks. NOTE: You need to assign at least three atoms for this to work - bonus points if you can explain why! All other uric acid atoms are given zero coordinates. SHELXL interprets this as "keep the geometry of the fragment the same, but superimpose it on the major component by fitting the (three) specified atoms". That's the plan at least! Let's run a round of refinement and see what happens. The view in ShelXle should look like this:
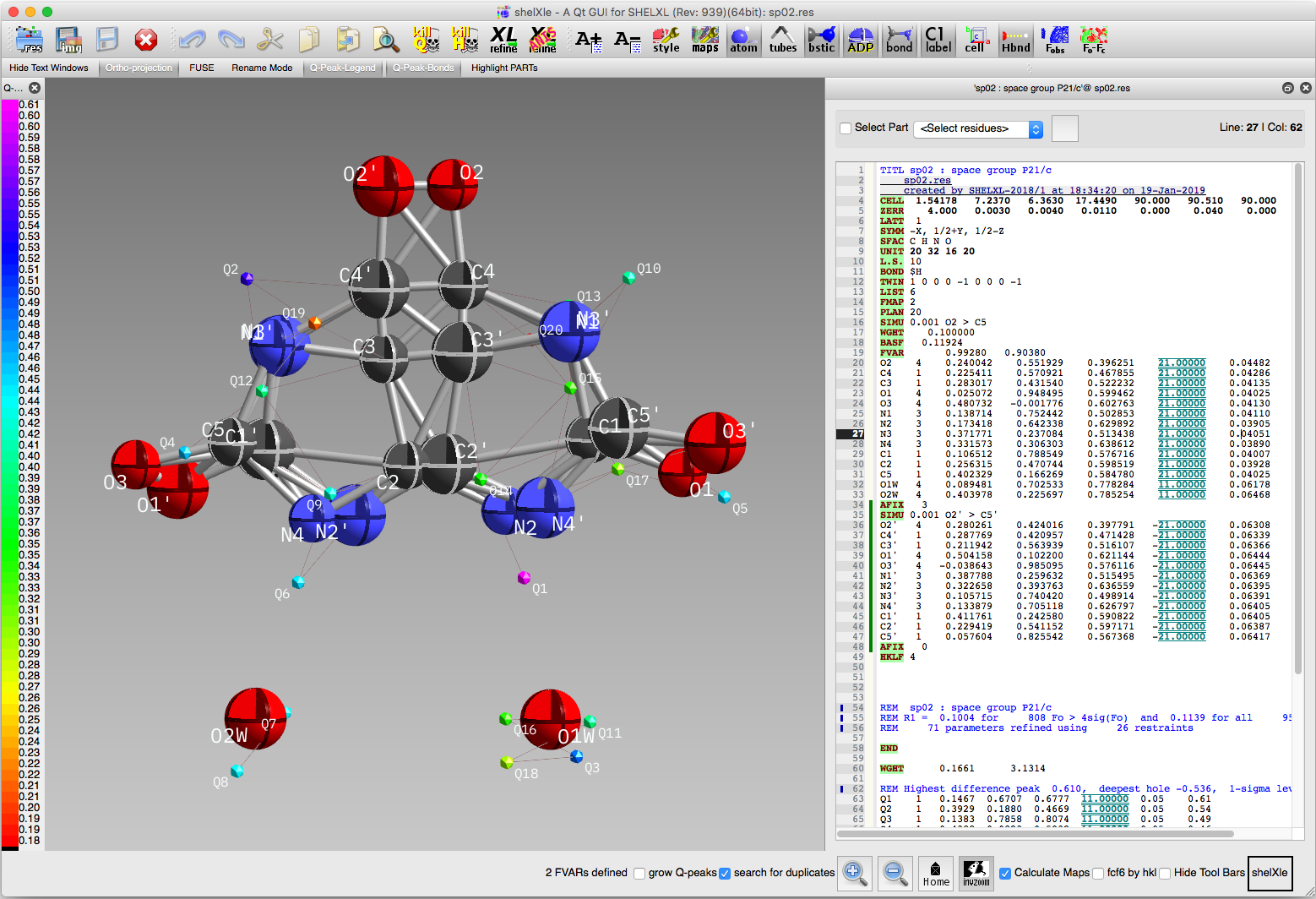
The superposition has worked as planned, but the structure still looks a bit
messy. Unrealistic 'bonds' have been drawn all over the place and the size of
the ellipsoids for atoms that overlap are too dissimilar. The first of these
problems will be fixed by assigning the major and minor disorder components to
separate PARTs, as in the sorbose
and whole-molecule disorder
tutorials. The rest will be fixed by imposing a more appropriate set of restraints
on the model prior to going anisotropic, adding hydrogens, and completing the
refinement.
1) Solve structure using SHELXT and ShelXle.
2) Include twinning using TWIN and BASF in SHELXL.
3) Add disorder using FRAG...FEND in SHELXL.
4) Add restraints/constraints to enable stable refinement.
... More advanced stuff (only possible because β is very close to 90°)...
5) Tie the major/minor disorder components using FVARs.
2) Include twinning using TWIN and BASF in SHELXL.
3) Add disorder using FRAG...FEND in SHELXL.
4) Add restraints/constraints to enable stable refinement.
... More advanced stuff (only possible because β is very close to 90°)...
5) Tie the major/minor disorder components using FVARs.
Return to the first page of this tutorial
Return to the main Tutorials page or to the main X-Ray Lab page
Return to the main Tutorials page or to the main X-Ray Lab page